Search Count: 51
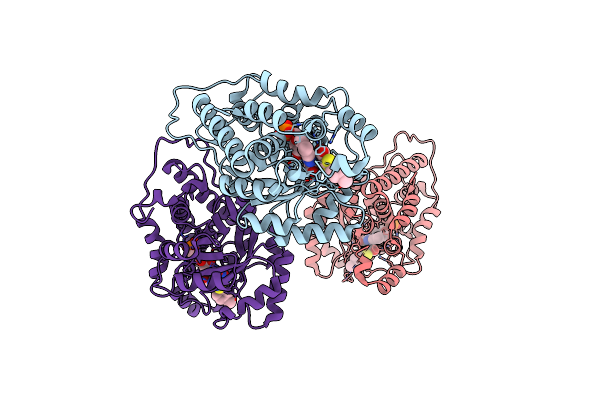 |
High Resolution Crystal Structure Of Rat Long Chain Hydroxy Acid Oxidase In Complex With The Inhibitor 4-Carboxy-5-[(4-Chiorophenyl)Sulfanyl]-1, 2, 3-Thiadiazole.
Organism: Rattus norvegicus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2012-03-07 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: FMN, HO6 |
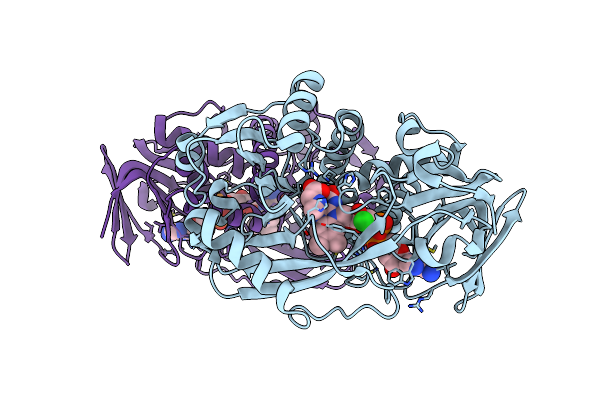 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, SAR |
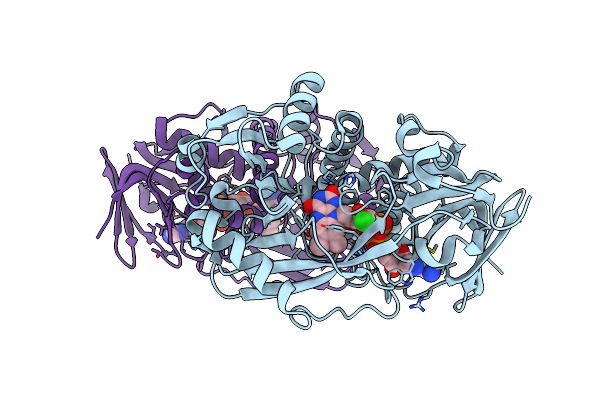 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
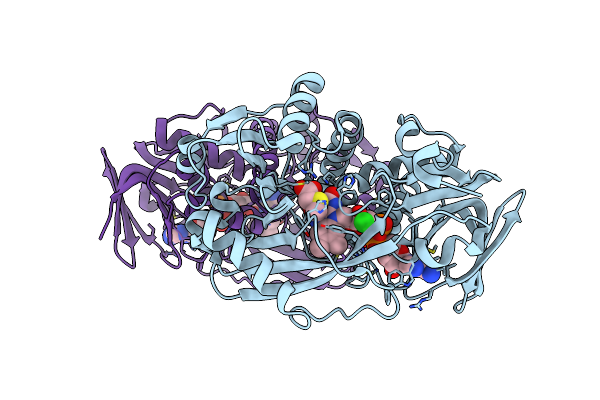 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: FAD, CL, MTG |
 |
Crystal Structure Of Glucose Oxidase For Space Group C2221 At 1.2 A Resolution
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, GOL, PEG, CL |
 |
Crystal Structure Of Glucose Oxidase For Space Group P3121 At 1.3 A Resolution.
Organism: Aspergillus niger
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-06-01 Classification: OXIDOREDUCTASE Ligands: NAG, FAD, CL |
 |
Crystal Structural Of Mutant Y305A In The Copper Amine Oxidase From Hansenula Polymorpha
Organism: Pichia angusta
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2010-08-25 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Crystal Structure Of Mutant Y305F Expressed In E. Coli In The Copper Amine Oxidase From Hansenula Polymorpha
Organism: Pichia angusta
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2010-08-25 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Crystal Structure Of Y305F Mutant Of The Copper Amine Oxidase From Hansenula Polymorpha Expressed In Yeast
Organism: Pichia angusta
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-08-25 Classification: OXIDOREDUCTASE Ligands: CU, PO4 |
 |
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of The Lys265Arg Phosphate-Crytsallized Mutant Of Monomeric Sarcosine Oxidase
Organism: Bacillus sp. b-0618
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL |
 |
Crystal Structure Of The Lys265Arg Peg-Crystallized Mutant Of Monomeric Sarcosine Oxidase
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-04-21 Classification: OXIDOREDUCTASE Ligands: FAD, CL, PO4 |
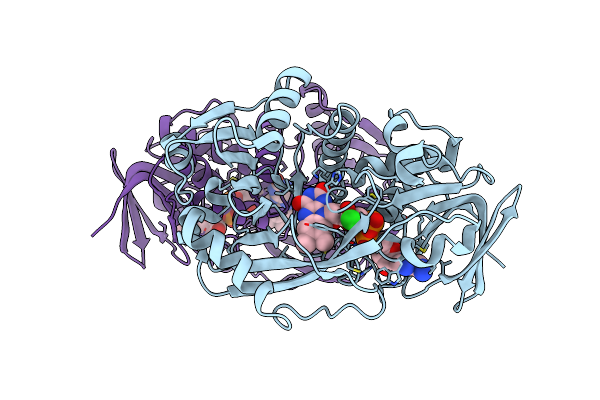 |
Crystal Structure Of R49K Mutant Of Monomeric Sarcosine Oxidase Crystallized In Peg As Precipitant
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2008-02-26 Classification: OXIDOREDUCTASE Ligands: CL, FAD |
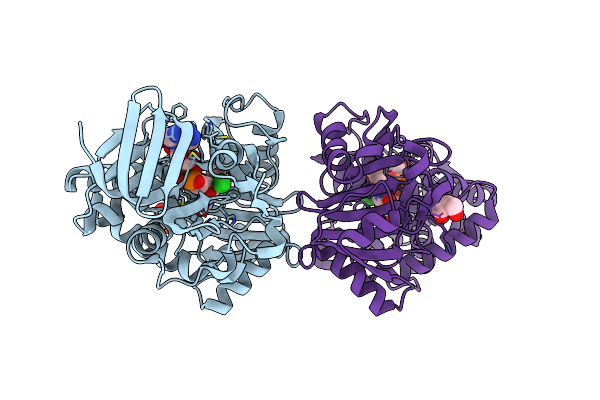 |
Crystal Structure Of R49K Mutant Of Monomeric Sarcosine Oxidase Crystallized In Phosphate As Precipitant
Organism: Bacillus sp.
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2008-02-26 Classification: OXIDOREDUCTASE Ligands: CL, FAD, GOL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2007-04-10 Classification: HYDROLASE Ligands: NAG |
 |
Human Thrombin Chimera With Human Residues 184A, 186, 186A, 186B, 186C And 222 Replaced By Murine Thrombin Equivalents.
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-04-10 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: 0G6, NAG |
 |
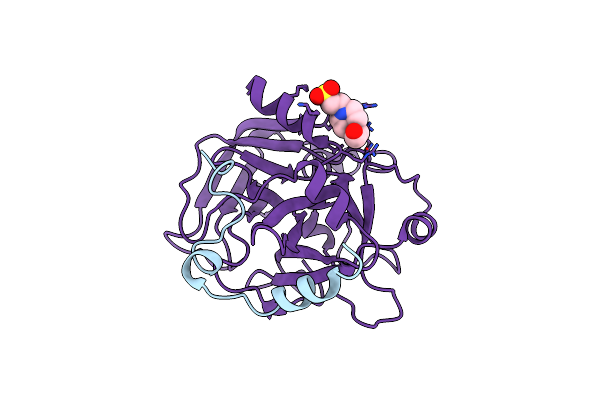
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2006-09-19 Classification: HYDROLASE |
 |
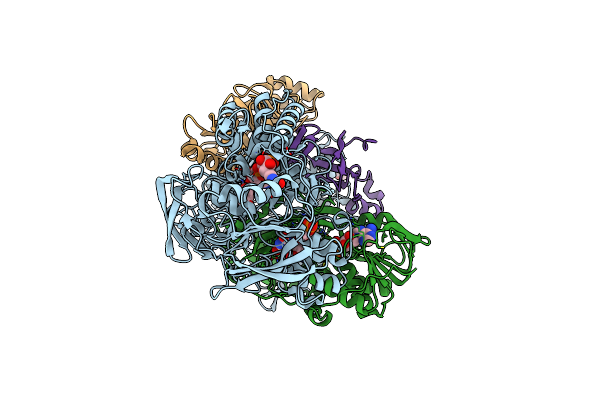
Crystal Structure Of The Slow Form Of Thrombin In A Self-Inhibited Conformation
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2006-09-12 Classification: HYDROLASE Ligands: EPE |
 |
Heteroteterameric Sarcosine: Structure Of A Diflavin Metaloenzyme At 1.85 A Resolution
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-08-08 Classification: OXIDOREDUCTASE Ligands: FOA, NAD, GOL, SO3, FAD, FMN, ZN |
 |
Heterotetrameric Sarcosine: Structure Of A Diflavin Metaloenzyme At 1.85 A Resolution
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-08-08 Classification: OXIDOREDUCTASE Ligands: NAD, FAD, FMN, FOA, ZN |

