Search Count: 1,470
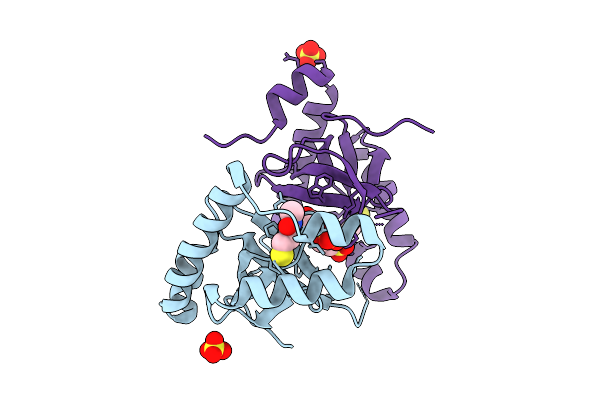 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: SO4, A1EF6, EPE |
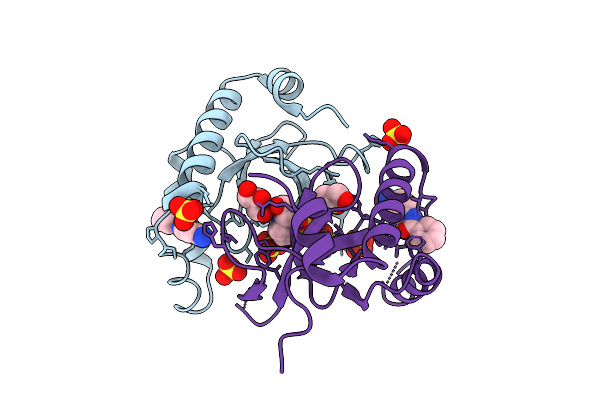 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: 6Y3, SO4, GOL, EPE |
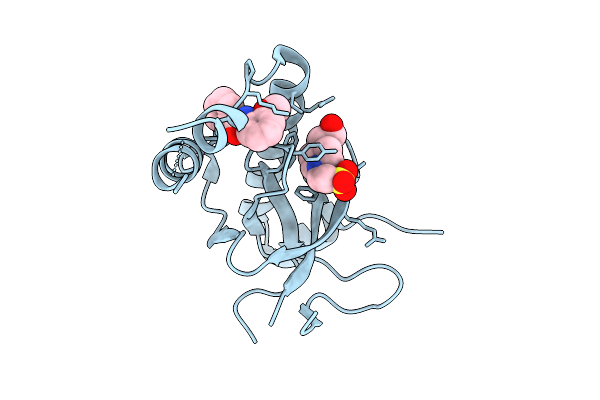 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-26 Classification: TRANSFERASE Ligands: EPE, A1EF7 |
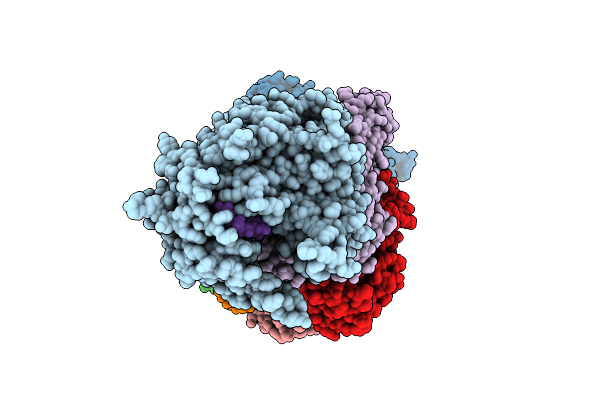 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: MN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Bacteroides fragilis
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: IMMUNE SYSTEM/RNA Ligands: ATP, SAM, MN |
 |
Organism: Natrinema sp. j7-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-26 Classification: PROTEIN FIBRIL |
 |
Organism: Human alphaherpesvirus 3
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: VIRUS |
 |
Crystal Structure Of The Bromodomain Of Human Brd9 In Complex With The Inhibitor Y22076
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: PROTEIN BINDING Ligands: A1EKN, DMS |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B74 |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B73 |
 |
Crystal Structure Of The Bromodomain Of Human Brd9 In Complex With The Inhibitor Y22024
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EF0, GOL, MLT |
 |
Crystal Structure Of The Bromodomain Of Human Brd9 In Complex With The Inhibitor Y22032
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EHA |
 |
Crystal Structure Of The Bromodomain Of Human Brd9 In Complex With The Inhibitor Y22077
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: PROTEIN BINDING Ligands: A1EFZ, EDO |
 |
Crystal Structure Of Arabidopsis Thaliana Ing1 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
 |
Crystal Structure Of Arabidopsis Thaliana Ing2 Phd Finger In Complex With An H3K4Me3 Peptide
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Release Date: 2025-11-05 Classification: GENE REGULATION Ligands: ZN |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |
 |
Organism: Salmonella enterica subsp. diarizonae
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: TOXIN |

