Search Count: 89
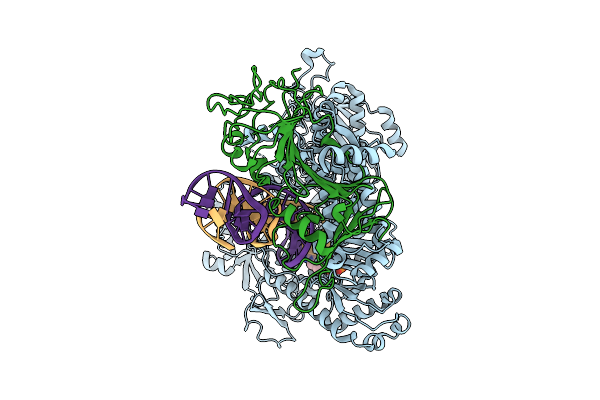 |
Herpes Simplex Virus Type 1 Polymerase Machinery In Complex With Duplex Dna, Acyclovir Triphosphate And Calcium Ions
Organism: Human herpesvirus 1 (strain 17), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-09-03 Classification: REPLICATION Ligands: CA, AVP |
 |
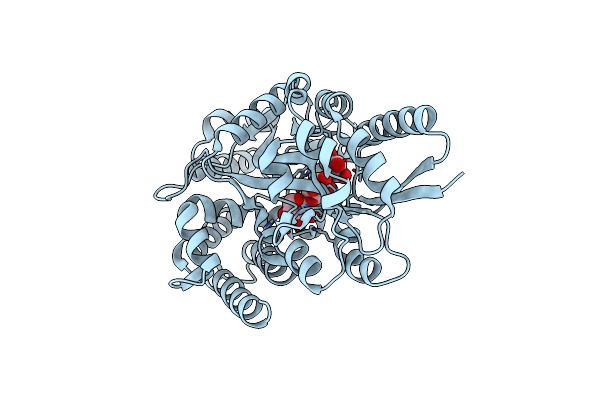
Crystal Structure Of Psts From Candidatus Pelagibacter Sp. Htcc7211 In Complex With Phosphate
Organism: Candidatus pelagibacter sp. htcc7211
Method: X-RAY DIFFRACTION Release Date: 2025-07-30 Classification: PROTEIN BINDING Ligands: PO4 |
 |
Organism: Isochrysis galbana
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, DD6, LHG, LMG, A86, LMU, SQD, DGD, BCR, SF4, PQN |
 |
The Substrate Binding Protein Of An Abc Transporter In Complex With Beta-1,4-Xylobiose
Organism: Vibrio sp. ea2
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: TRANSPORT PROTEIN Ligands: GOL |
 |
The Substrate Binding Protein Of An Abc Transporter In Complex With Beta-1,3-Xylobiose
Organism: Vibrio sp. ea2
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: TRANSPORT PROTEIN |
 |
The Substrate Binding Protein Of An Abc Transporter In Complex With Beta-1,3-Xylotriose
Organism: Vibrio sp. ea2
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: TRANSPORT PROTEIN |
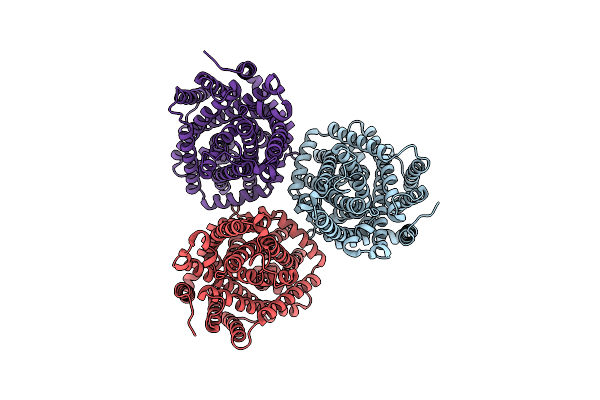 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT |
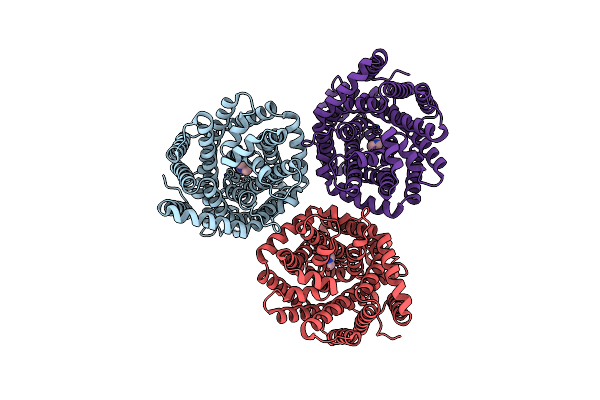 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT Ligands: KEN |
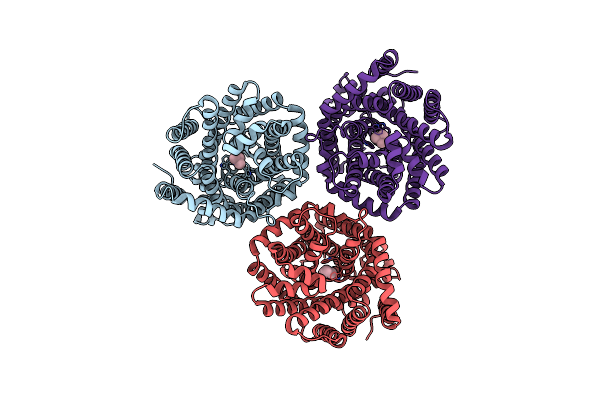 |
Organism: Myroides profundi
Method: ELECTRON MICROSCOPY Release Date: 2024-11-06 Classification: ELECTRON TRANSPORT Ligands: KEN |
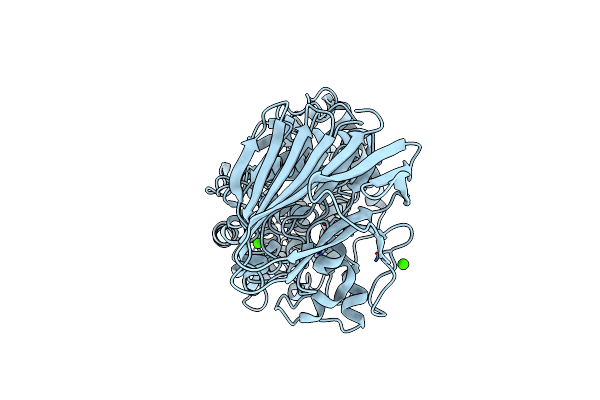 |
Organism: Spirosoma fluviale
Method: X-RAY DIFFRACTION Release Date: 2024-09-04 Classification: LYASE Ligands: MN, CA |
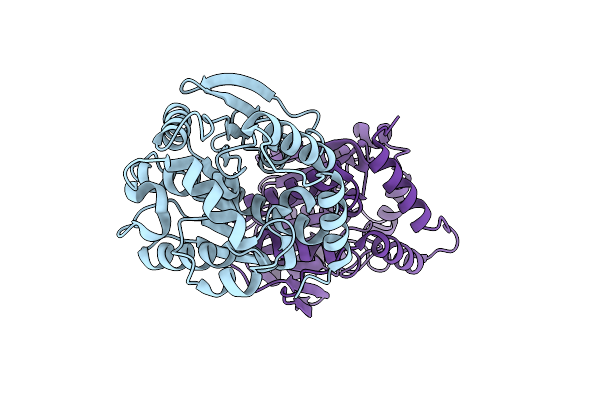 |
Organism: Paracoccus kondratievae
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2024-07-31 Classification: HYDROLASE |
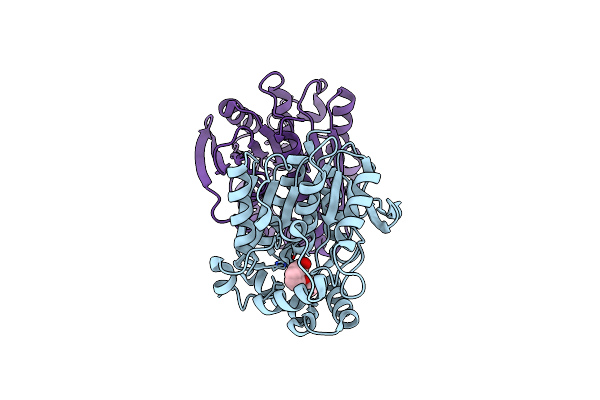 |
Organism: Paracoccus kondratievae
Method: X-RAY DIFFRACTION Resolution:1.76 Å Release Date: 2024-07-31 Classification: HYDROLASE Ligands: A1D6Y |
 |
Organism: Chroomonas placoidea
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: PHOTOSYNTHESIS Ligands: OEX, FE2, BCT, CL, CLA, PHO, 8CT, PL9, LHG, KC2, II0, IHT, SQD, LMG, DGD, HEM |
 |
Organism: Symbiodinium sp.
Method: ELECTRON MICROSCOPY Release Date: 2024-05-15 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, UIX, DD6, SQD, PID, LMG, LHG, DGD, LMU, PQN, BCR, SF4 |
 |
Crystal Structure Of Urta From Prochlorococcus Marinus Str. Mit 9313 In Complex With Urea And Calcium
Organism: Prochlorococcus marinus str. mit 9313
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2023-11-22 Classification: TRANSPORT PROTEIN Ligands: URE, CA |
 |
Organism: Acinetobacter bereziniae niph 3
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2023-10-04 Classification: LYASE Ligands: ZN, LNI |
 |
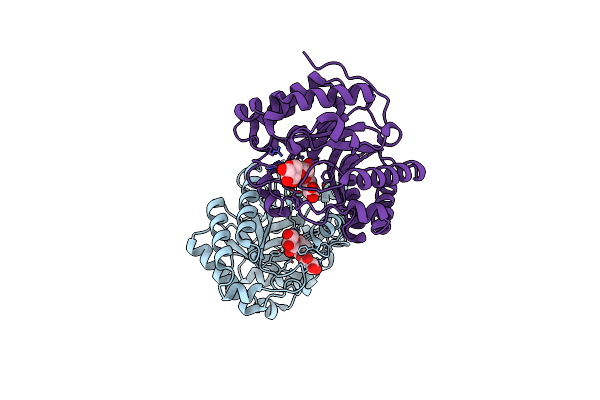
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-10-04 Classification: LYASE Ligands: MN, LNI |
 |
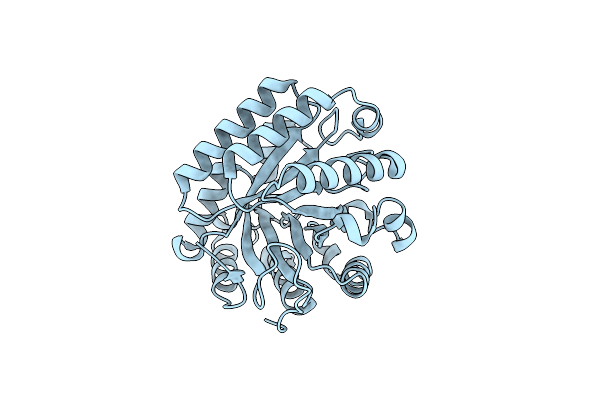
Organism: Algibacter sp. l4_22
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-05-17 Classification: HYDROLASE |
 |
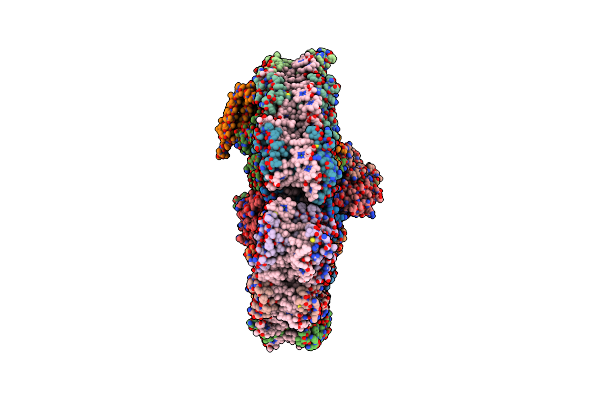
Organism: Algibacter sp. l4_22
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2023-04-26 Classification: HYDROLASE |
 |
Organism: Chroomonas placoidea
Method: ELECTRON MICROSCOPY Release Date: 2023-04-12 Classification: PHOTOSYNTHESIS Ligands: CLA, KC2, II0, II3, IHT, LMG, LHG, SQD, 8CT, LMU, PQN, SF4, DGD |

