Search Count: 21
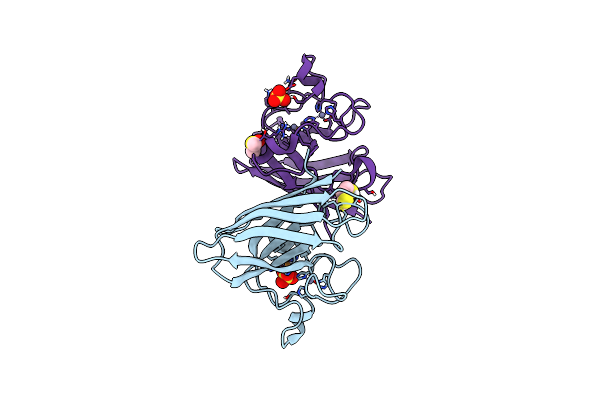 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: CU, ZN, SO4, UDI, DMS |
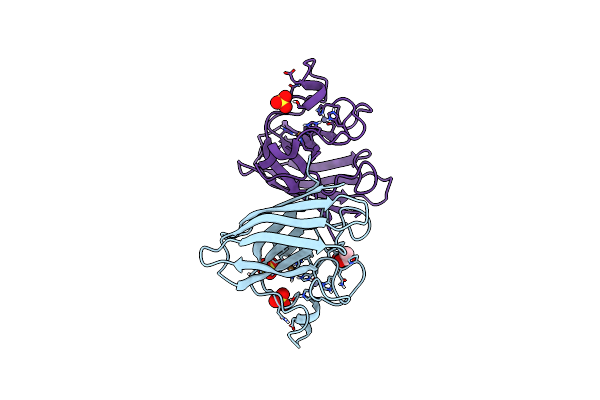 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2023-12-13 Classification: OXIDOREDUCTASE Ligands: ACT, ZN, CU, SO4 |
 |
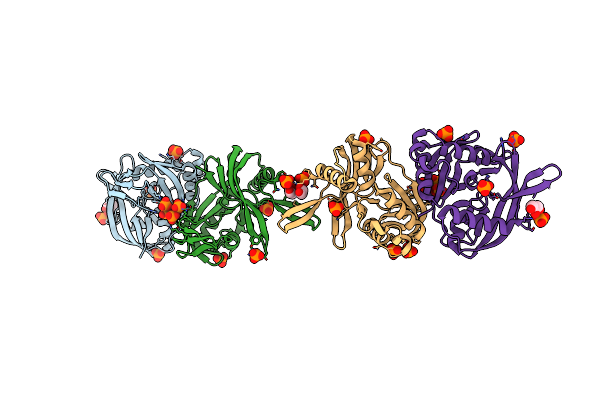
Crystal Structure Of Canine Coronavirus Main Protease In Complex With Gc376
Organism: Canine coronavirus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-05-31 Classification: VIRAL PROTEIN/INHIBITOR Ligands: K36 |
 |
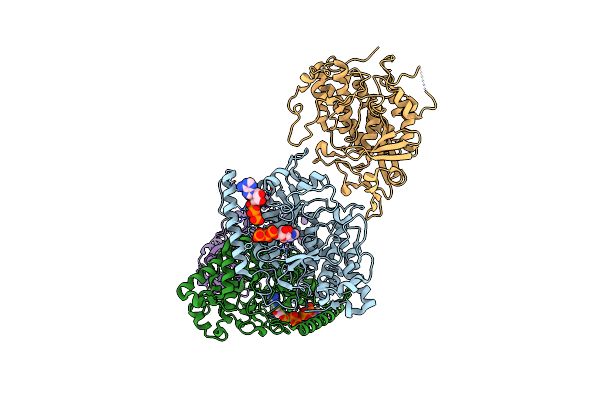
Organism: Methanocaldococcus jannaschii dsm 2661
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2018-09-12 Classification: HYDROLASE Ligands: GOL, PO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.30 Å Release Date: 2018-04-25 Classification: TRANSFERASE Ligands: ATP |
 |
Organism: Hydra vulgaris
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2018-04-11 Classification: TRANSFERASE Ligands: NAG |
 |
Organism: Hydra vulgaris
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2018-04-11 Classification: TRANSFERASE Ligands: NAG, MBN, ADN |
 |
Organism: Danio rerio
Method: X-RAY DIFFRACTION Resolution:3.45 Å Release Date: 2018-04-11 Classification: TRANSFERASE |
 |
Organism: Homo sapiens, Danio rerio
Method: X-RAY DIFFRACTION Resolution:3.55 Å Release Date: 2018-04-11 Classification: TRANSFERASE Ligands: ATP |
 |
Crystal Structure Of Ketosteroid Isomerase Variant M105A From Pseudomonos Putida
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2011-08-03 Classification: ISOMERASE |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2011-07-06 Classification: HYDROLASE Ligands: 5P5 |
 |
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2011-03-23 Classification: LYASE Ligands: 3CO, GOL |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Beta-E56Q From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2011-03-23 Classification: LYASE Ligands: GOL, 3CO |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Beta-H71L From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-03-23 Classification: LYASE Ligands: 3CO |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Alpha-E168Q From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-03-23 Classification: LYASE Ligands: GOL, 3CO |
 |
Crystal Structure Of Co-Type Nitrile Hydratase Beta-Y215F From Pseudomonas Putida.
Organism: Pseudomonas putida
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2011-03-23 Classification: LYASE Ligands: GOL, 3CO |
 |
Structure Of The Aml1-Eto Nervy Domain - Pka(Riia) Complex And Its Contribution To Aml1-Eto Activity
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2010-10-20 Classification: PROTEIN BINDING |
 |
Crystal Structure Of A Putative Metal-Dependent Phosphoesterase (Bad_1165) From Bifidobacterium Adolescentis Atcc 15703 At 1.94 A Resolution
Organism: Bifidobacterium adolescentis
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: AMP, ZN, FE, PO4, EDO |
 |
Crystal Structure Of A Putative Metal-Dependent Phosphoesterase (Bad_1165) From Bifidobacterium Adolescentis Atcc 15703 At 2.40 A Resolution
Organism: Bifidobacterium adolescentis atcc 15703
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2008-09-02 Classification: HYDROLASE Ligands: ZN, FE, PO4, ACT |

