Search Count: 28
 |
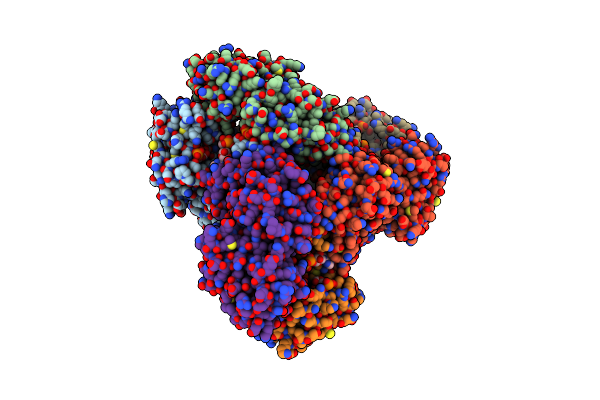
Crystal Strcuture Of Human Phosphoribosyl Pyrophosphate Synthetase2 (Prps2) In Complex With Ligands
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN Ligands: APC, HSX, CD, PO4 |
 |
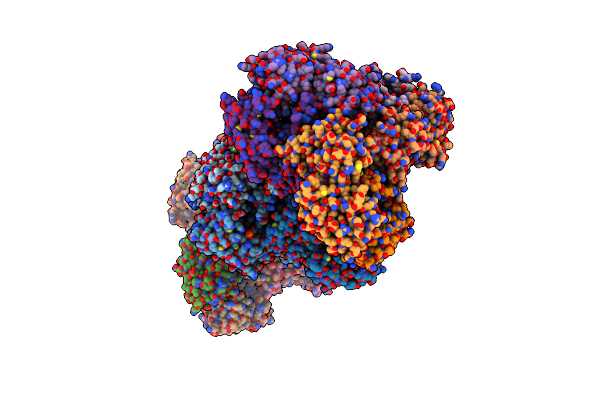
Crystal Strcture Of Human Phosphoribosyl Pyrophosphate Synthetase 1 (Prps1) In Complex With Gdp
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN Ligands: APC, HSX, GDP, PO4 |
 |
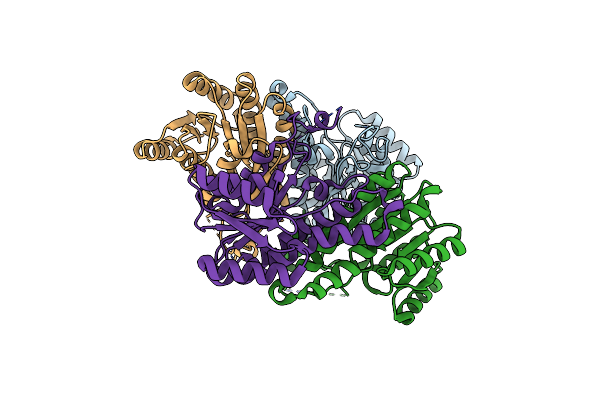
Crystal Structure Of Human Phosphoribosyl Pyrophosphate Synthetase 1(Prps1) Chimera Swapped With Three Residues From Prps2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2025-03-19 Classification: BIOSYNTHETIC PROTEIN Ligands: HSX |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of 3-Oxoacyl-Acp Reductase Fabg In Complex With Nadph From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP |
 |
Crystal Structure Of 3-Oxoacyl-Acp Reductase Fabg In Complex With Nadp+ And 3-Keto-Octanoyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.83 Å Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, UHC |
 |
Crystal Structure Of 3-Oxoacyl-Acp Reductase Fabg In Complex With Nadp+ And 3-Keto-Octanoyl-Acp From Helicobacter Pylori In An Inactive Form That Priors The Acyl Substrate Delivery
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, UHC, PN7 |
 |
Crystal Structure Of 3-Oxoacyl-Acp Reductase Fabg In Complex With Nadp+ And 3-Keto-Hexanoyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN Ligands: NAP, UHC |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of Enoyl-Acp Reductase Fabi In Complex With Nadh From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN Ligands: NAD |
 |
Crystal Structure Of Enoyl-Acp Reductase Fabi In Complex With Nad+ And Crotonyl-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.41 Å Release Date: 2023-11-15 Classification: BIOSYNTHETIC PROTEIN Ligands: NAD, PSR |
 |
Crystal Structure Of The Dioxygenase Cctet From Coprinopsis Cinereain In Complex With Mn(Ii) And N-Oxalylglycine
Organism: Coprinopsis cinerea (strain okayama-7 / 130 / atcc mya-4618 / fgsc 9003)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-03-30 Classification: DNA BINDING PROTEIN Ligands: MN, OGA |
 |
Crystal Structure Of The Dioxygenase Cctet From Coprinopsis Cinereain Bound To 12Bp N6-Methyldeoxyadenine (6Ma) Containing Duplex Dna
Organism: Coprinopsis cinerea, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-03-30 Classification: DNA BINDING PROTEIN/DNA Ligands: MN, OGA |
 |
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-12-01 Classification: FLAVOPROTEIN Ligands: SF4, FMN, CL |
 |
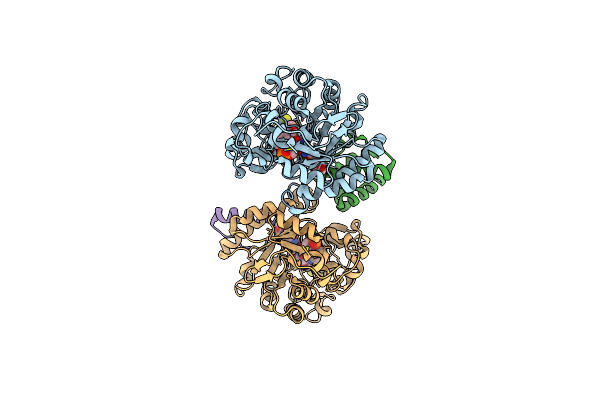
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Holo-Acp
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-12-01 Classification: FLAVOPROTEIN Ligands: FMN, SF4, PN7 |
 |
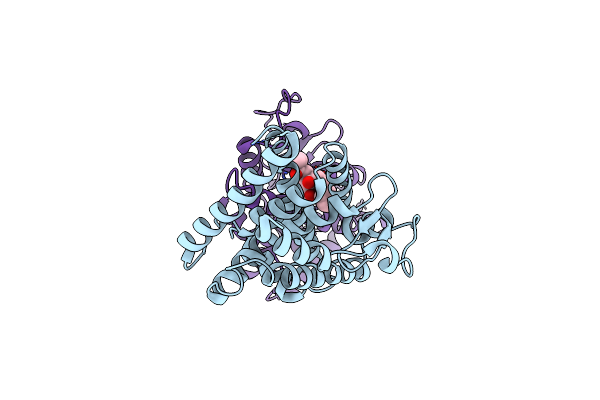
Crystal Structure Of Dehydrogenase/Isomerase Fabx From Helicobacter Pylori In Complex With Octanoyl-Acp
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.31 Å Release Date: 2021-12-01 Classification: FLAVOPROTEIN Ligands: FMN, SF4, 66S |
 |
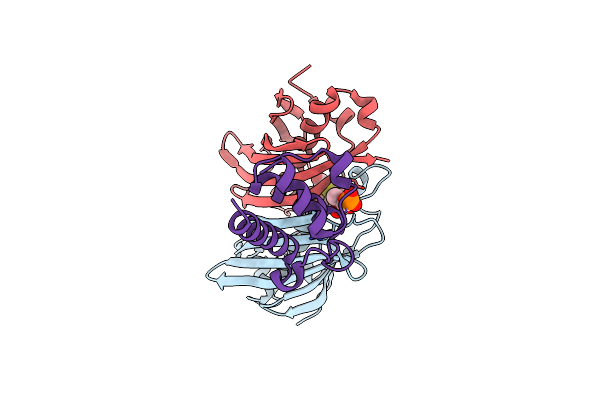
Crystal Structure Analysis Of The Csn-B-Bounded Nur77 Ligand Binding Domain
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.45 Å Release Date: 2020-10-14 Classification: TRANSCRIPTION Ligands: E4L |
 |
Crystal Structure Of (3R)-Hydroxyacyl-Acyl Carrier Protein Dehydratase(Fabz) In Complex With Holo-Acp From Helicobacter Pylori
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2016-11-02 Classification: LYASE/BIOSYNTHETIC PROTEIN Ligands: CIT, PNS |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2015-09-09 Classification: TRANSCRIPTION Ligands: GOL, 3MJ |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-09-09 Classification: TRANSCRIPTION Ligands: GOL, 3N0 |

