Search Count: 16
 |
Structure Of Trehalose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside-1,2-Lyase Al1
Organism: Alistipes
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-06-19 Classification: ISOMERASE Ligands: CO, ACT |
 |
Organism: Alistipes
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2024-06-19 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Alistipes
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2024-06-19 Classification: HYDROLASE Ligands: MG |
 |
Organism: Alistipes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2024-06-12 Classification: ISOMERASE Ligands: CO, GOL, ACT |
 |
Structure Of Glucose Bound Alistipes Sp. 3-Keto-Beta-Glucopyranoside-1,2-Lyase Al1
Organism: Alistipes
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2024-06-12 Classification: ISOMERASE Ligands: MLI, GLC, CO |
 |
Structure Of 3K-Glch Bound Bacteroides Thetaiotaomicron 3-Keto-Beta-Glucopyranoside-1,2-Lyase Bt1
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2024-06-12 Classification: ISOMERASE Ligands: UC6, CO, PO4 |
 |
Structure Of Bacteroides Thetaiotaomicron 3-Keto-2-Hydroxy-Glucal-Hydratase Bt2
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-06-12 Classification: ISOMERASE Ligands: K |
 |
Structure Of Glucose Bound Bacteroides Thetaiotaomicron 3-Keto-2-Hydroxy-Glucal-Hydratase Bt2
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-06-12 Classification: ISOMERASE Ligands: GLC, K |
 |
Organism: Alistipes
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-06-12 Classification: ISOMERASE Ligands: NAD |
 |
Organism: Collinsella tanakaei
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2024-06-12 Classification: OXIDOREDUCTASE Ligands: NAD, SO4, TRS |
 |
Human Pancreatic Alpha-Amylase In Complex With The Mechanism Based Inactivator Glucosyl Epi-Cyclophellitol
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.23 Å Release Date: 2016-07-06 Classification: HYDROLASE/HYDROLASE inhibitor Ligands: 5QP, CL, CA |
 |
Organism: Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158), Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:3.40 Å Release Date: 2016-03-30 Classification: METAL BINDING PROTEIN Ligands: C2E, XP5, 3PE |
 |
Organism: Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158), Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2016-03-09 Classification: MEMBRANE PROTEIN Ligands: PLC, 43Y, C2E, LDA, MG, 660 |
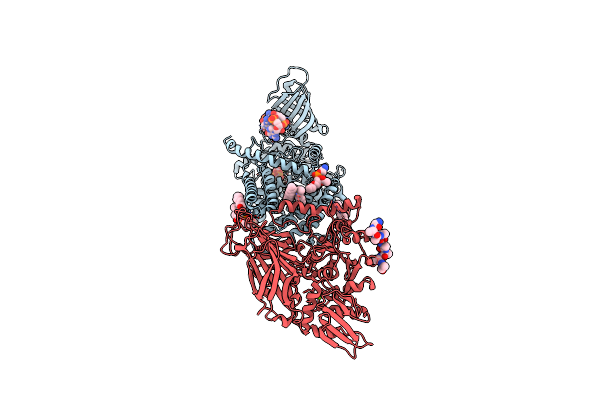 |
Organism: Rhodobacter sphaeroides (strain atcc 17023 / 2.4.1 / ncib 8253 / dsm 158), Rhodobacter sphaeroides
Method: X-RAY DIFFRACTION Resolution:2.94 Å Release Date: 2016-03-09 Classification: METAL BINDING PROTEIN Ligands: C2E, UDP, MG, PLC, 3PE, UND |
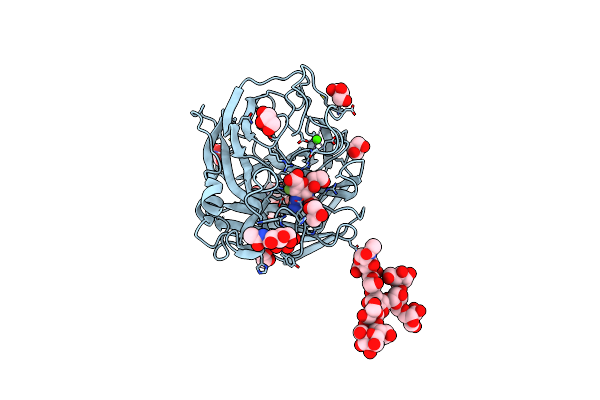 |
Influenza Virus Neuraminidase Subtype N9 (Tern) Complexed With 2,3-Dif Guanidino-Neu5Ac2En Inhibitor
Organism: Influenza a virus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2013-05-01 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: NAG, GOL, ZGE, CA |
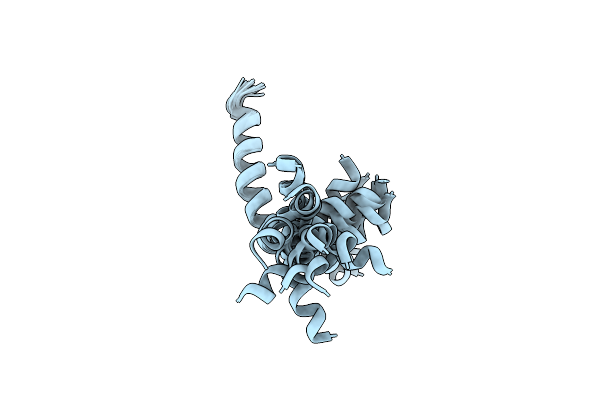 |
Solution Structure Of Cb1A, A Novel Anticancer Peptide Derived From Natural Antimicrobial Peptide Cecropin B
Organism: Hyalophora cecropia
Method: SOLUTION NMR Release Date: 2006-11-18 Classification: DE NOVO PROTEIN, LIPID BINDING PROTEIN |

