Search Count: 322
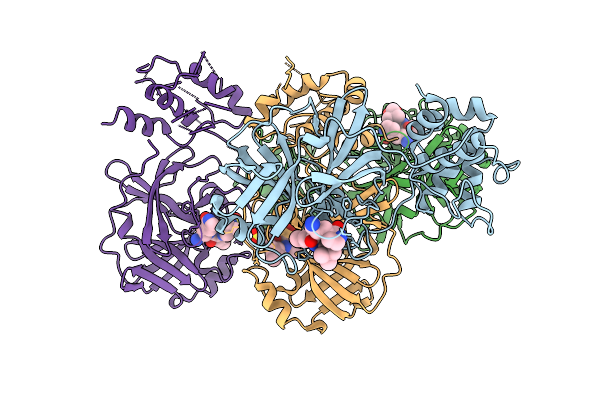 |
Organism: Canine coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7M |
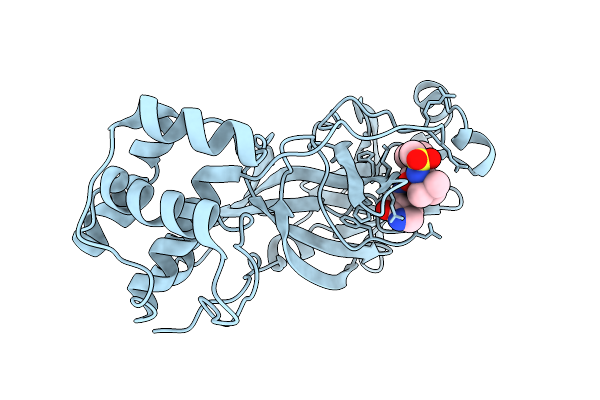 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7M |
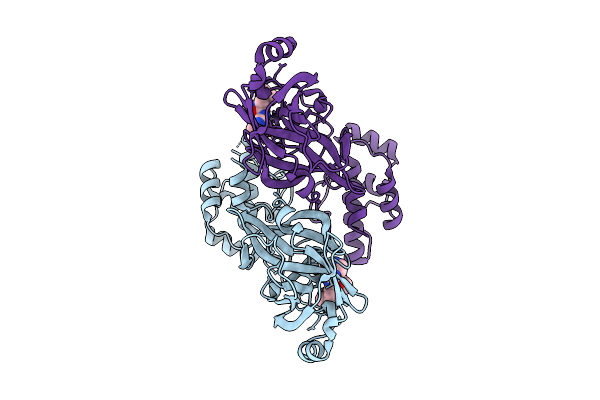 |
Organism: Avian infectious bronchitis virus (strain beaudette)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7M |
 |
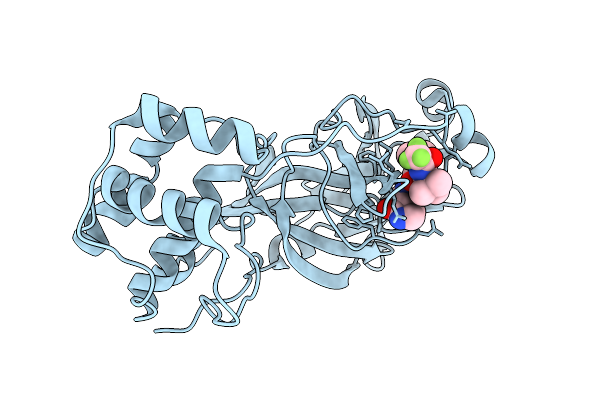
Crystal Structure Of Ccov-Hupn-2018 3Cl Protease (3Clpro) In Complex With Compound 8
Organism: Canine coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EPZ |
 |
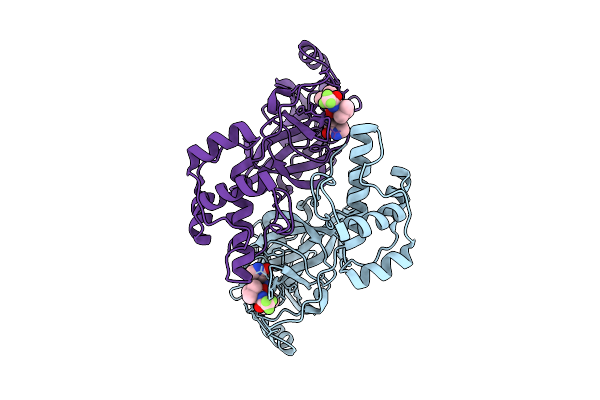
Crystal Structure Of Sars-Cov-2 3Cl Protease (3Clpro) In Complex With Compound 8
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EPZ |
 |
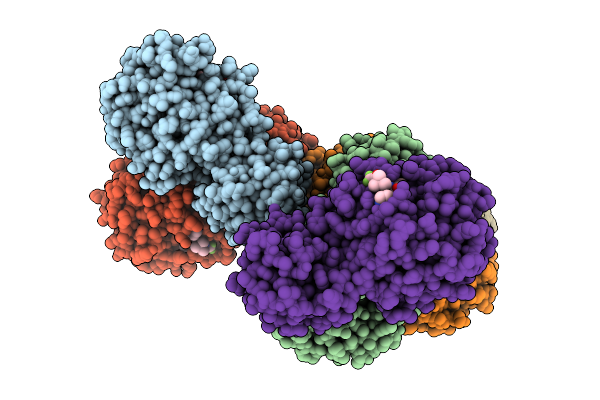
Organism: Avian infectious bronchitis virus (strain beaudette)
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EPZ |
 |
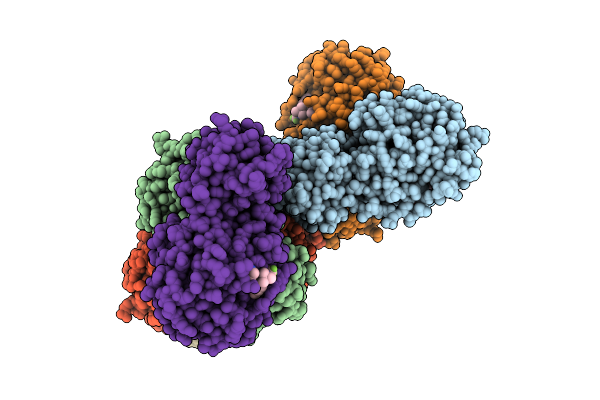
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 8
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1EPZ |
 |
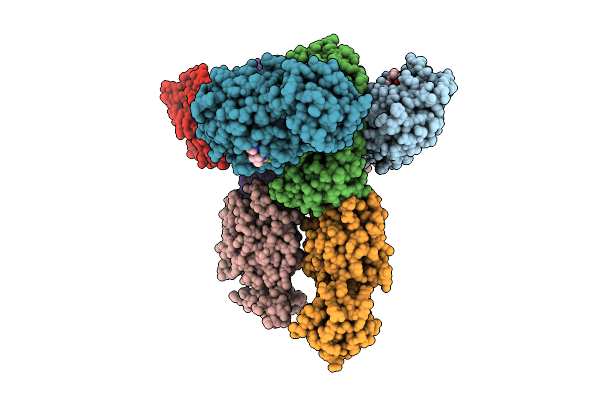
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Nirmatrelvir
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: 4WI |
 |
Crystal Of Ccov-Hupn-2018 3Cl Protease (3Clpro) In Complex With Nirmatrelvir
Organism: Canine coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: 4WI |
 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 1
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7G |
 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 3
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7N |
 |
Crystal Structure Of Pdcov 3Cl Protease (3Clpro) In Complex With Compound 6
Organism: Porcine deltacoronavirus
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7M |
 |
Organism: Canine coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-12-24 Classification: VIRAL PROTEIN Ligands: A1D7N |
 |
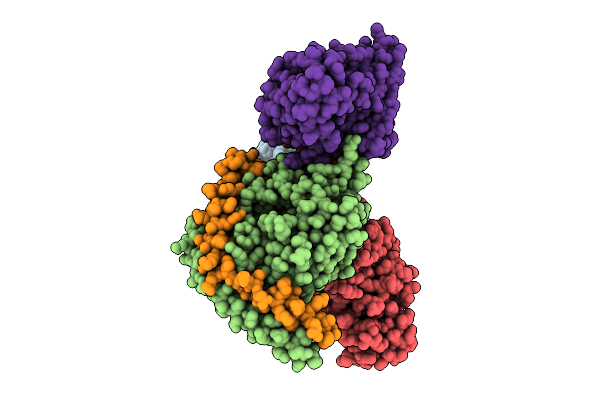
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
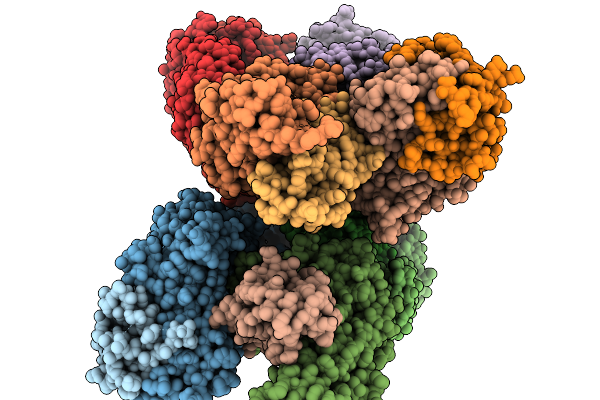
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: NIO |
 |
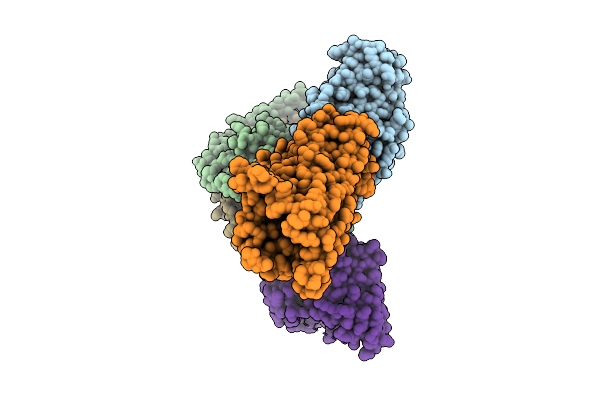
Organism: Homo sapiens, Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: IMMUNE SYSTEM Ligands: GOL, PEG |
 |
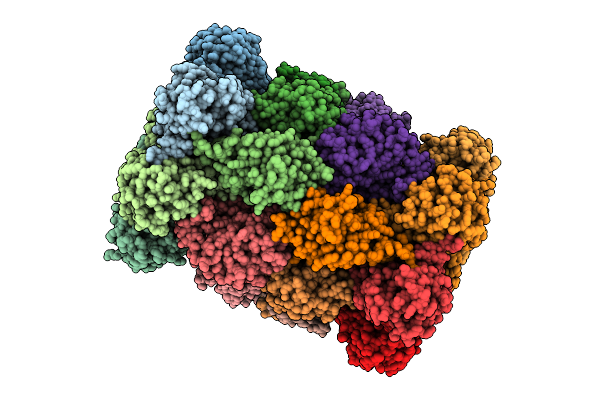
Organism: Homo sapiens, Vicugna pacos
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: PEPTIDE BINDING PROTEIN |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B74 |
 |
Organism: Plasmodium falciparum 3d7
Method: ELECTRON MICROSCOPY Release Date: 2025-11-12 Classification: CYTOSOLIC PROTEIN Ligands: A1B73 |
 |
Organism: Homo sapiens, Pyrococcus abyssi
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: MEMBRANE PROTEIN Ligands: 5YM, A1EM1, 1DO, A1EM3, A1EQ8, A1EM2 |

