Search Count: 20
 |
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2024-06-26 Classification: TRANSCRIPTION Ligands: XDO |
 |
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-06-26 Classification: TRANSCRIPTION Ligands: ACT, FO6 |
 |
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-03-02 Classification: TRANSCRIPTION Ligands: U5W |
 |
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2022-03-02 Classification: TRANSCRIPTION Ligands: U5W |
 |
Organism: Escherichia coli, Homo sapiens, Human papillomavirus type 16
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2019-09-04 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli, Human papillomavirus type 18, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2019-09-04 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Organism: Escherichia coli (strain k12), Human papillomavirus type 49, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2019-09-04 Classification: VIRAL PROTEIN Ligands: ZN, PEG |
 |
Organism: Escherichia coli (strain k12), Homo sapiens, Human papillomavirus type 16
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2019-08-21 Classification: VIRAL PROTEIN Ligands: ZN |
 |
Crystal Structure Of The 43K Atpase Domain Of Escherichia Coli Gyrase B In Complex With An Aminocoumarin
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2019-04-10 Classification: ISOMERASE Ligands: PO4, PG4, CL, NA, K, BHW |
 |
Crystal Structure Of The 43K Atpase Domain Of Thermus Thermophilus Gyrase B In Complex With An Aminocoumarin
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2019-04-10 Classification: ISOMERASE Ligands: BHW, IMD |
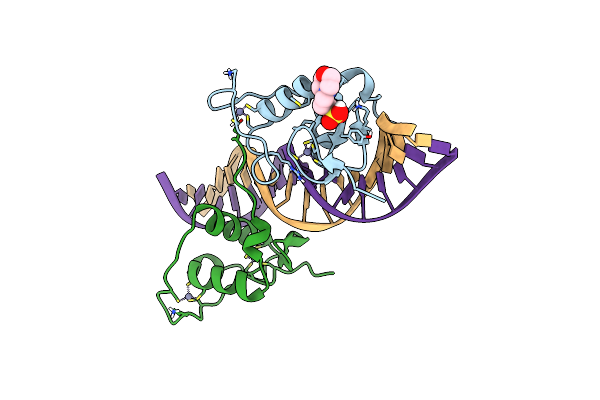 |
Crystal Structure Of The Human Retinoid X Receptor Dna-Binding Domain Bound To The Human Mep Dr1 Response Element, Ph 7.0
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2018-12-05 Classification: TRANSCRIPTION Ligands: ZN, MPO |
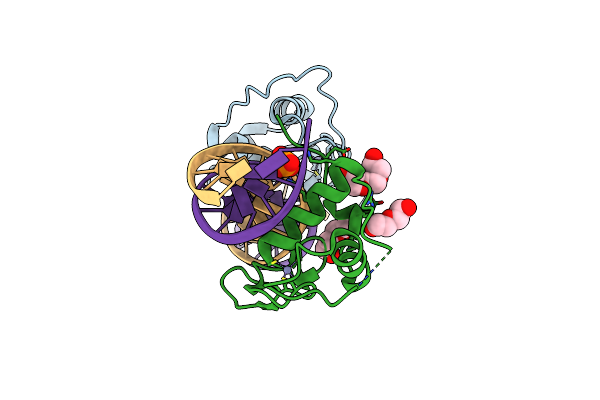 |
Crystal Structure Of The Human Retinoid X Receptor Dna-Binding Domain Bound To The Human Mep Dr1 Response Element, Ph 4.2
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2018-12-05 Classification: TRANSCRIPTION Ligands: ZN, PO4, PGE, PEG |
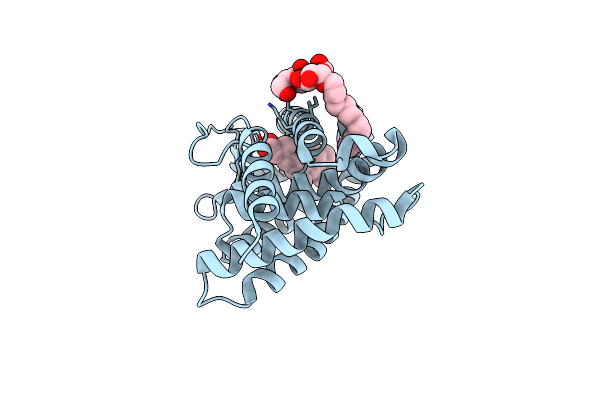 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.69 Å Release Date: 2017-09-20 Classification: NUCLEAR RECEPTOR Ligands: 9CR, LMU, CL |
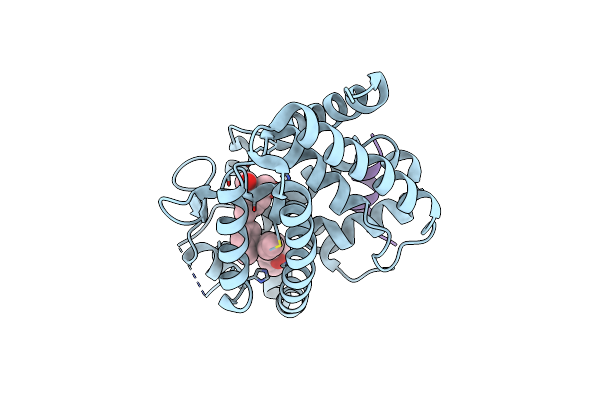 |
Structural Basis For The Accommodation Of Bis- And Tris-Aromatic Derivatives In Vitamin D Nuclear Receptor
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-09-26 Classification: transcription/transcription inhibitor Ligands: 0VK |
 |
Structural Basis For The Accommodation Of Bis- And Tris-Aromatic Derivatives In Vitamin D Nuclear Receptor
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2012-09-26 Classification: transcription/transcription inhibitor Ligands: 0VO |
 |
Structural Basis For The Accommodation Of Bis- And Tris-Aromatic Derivatives In Vitamin D Nuclear Receptor
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-09-26 Classification: transcription/transcription inhibitor Ligands: 0VP |
 |
Structural Basis For The Accommodation Of Bis- And Tris-Aromatic Derivatives In Vitamin D Nuclear Receptor
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-09-26 Classification: transcription/transcription inhibitor Ligands: 484 |
 |
Structural Basis For The Accommodation Of Bis- And Tris-Aromatic Derivatives In Vitamin D Nuclear Receptor
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2012-09-26 Classification: transcription/transcription inhibitor Ligands: 0VP |
 |
Structural Basis For The Accommodation Of Bis- And Tris-Aromatic Derivatives In Vitamin D Nuclear Receptor
Organism: Danio rerio, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2012-09-26 Classification: transcription/transcription inhibitor Ligands: 0VQ |
 |
Structural Basis For The Accommodation Of Bis- And Tris-Aromatic Derivatives In Vitamin D Nuclear Receptor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-09-26 Classification: transcription/transcription inhibitor Ligands: 0VQ |

