Search Count: 152
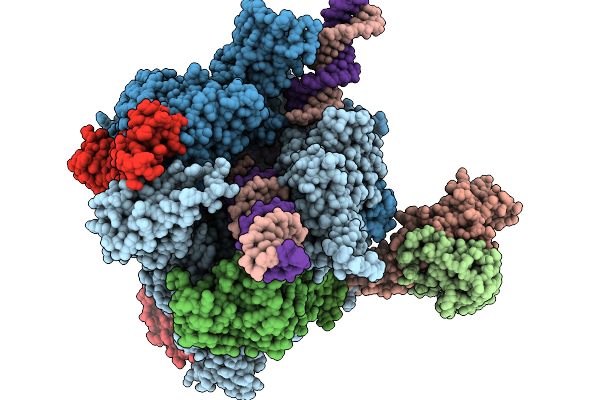 |
Structure Of A Native Drosophila Melanogaster Pol Ii Elongation Complex With A Well-Defined Rpb4/Rpb7 Stalk
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Resolution:3.44 Å Release Date: 2026-01-07 Classification: TRANSCRIPTION Ligands: ZN |
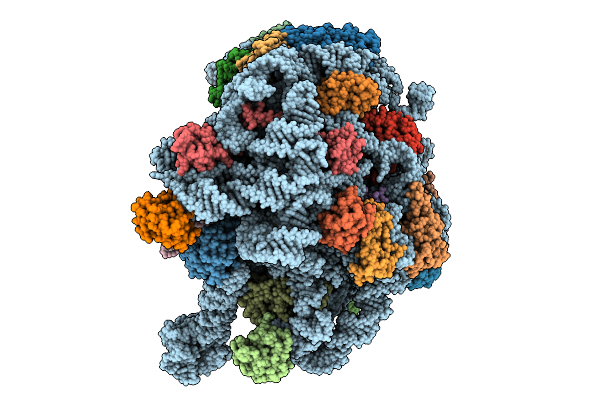 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
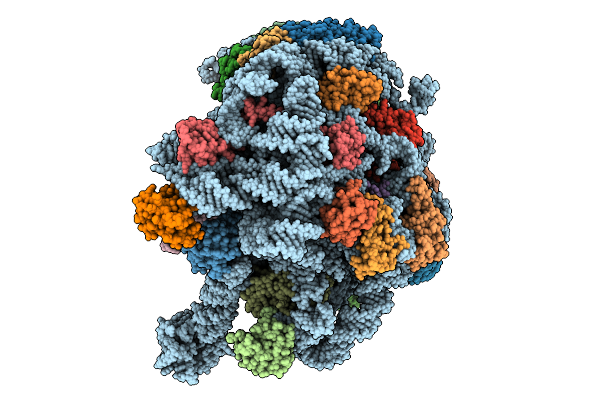 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, K, ZN |
 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME Ligands: MG, ZN |
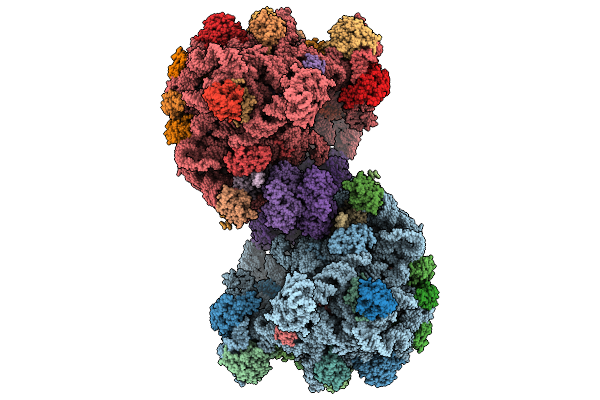 |
Organism: Methanosarcina acetivorans
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: RIBOSOME |
 |
Organism: Sars-cov-2 pseudovirus
Method: X-RAY DIFFRACTION Resolution:2.38 Å Release Date: 2025-09-17 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JLH, CL, IMD |
 |
Crystal Structure Of Ddb1-Crbn-Alv1 Complex Bound To Triple Znf Of Helios (Ikzf2 Zf1-3)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:4.11 Å Release Date: 2025-09-03 Classification: LIGASE Ligands: RN9, ZN |
 |
Organism: Severe acute respiratory syndrome coronavirus
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2025-08-27 Classification: VIRAL PROTEIN Ligands: ZN, SAH, A1JDF, CL, EDO, IMD, IPA, SO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.20 Å Release Date: 2025-08-20 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:3.10 Å Release Date: 2025-08-20 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-08-20 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LIGASE Ligands: ZN, A1BC8 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-08-06 Classification: LIGASE Ligands: ZN, A1BC8 |
 |
Organism: Synthetic construct, Xenopus laevis, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN/DNA |
 |
Organism: Synthetic construct, Xenopus laevis, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN/DNA |
 |
2.88 A S.Cerevisiae Chd1[L886G/L889G/L891G]-Nucleosome 2:1 Complex With Dna-Binding Domain
Organism: Xenopus laevis, Saccharomyces cerevisiae, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-06-11 Classification: DNA BINDING PROTEIN/DNA |
 |
Structure Of A Native Drosophila Melanogaster Pol Ii Elongation Complex Without Rpb4/Rpb7 Stalk
Organism: Drosophila melanogaster
Method: ELECTRON MICROSCOPY Resolution:3.37 Å Release Date: 2025-05-14 Classification: TRANSCRIPTION Ligands: ZN |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: SIGNALING PROTEIN Ligands: ATP |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: SIGNALING PROTEIN Ligands: ATP |
 |
Organism: Homo sapiens, Bos taurus
Method: ELECTRON MICROSCOPY Release Date: 2025-04-23 Classification: SIGNALING PROTEIN Ligands: ATP |

