Search Count: 49
 |
Organism: Dipsastraea favus
Method: X-RAY DIFFRACTION Release Date: 2025-08-27 Classification: LUMINESCENT PROTEIN |
 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Double Mutant Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: CA, A1H0D, LI |
 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Single Mutant Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: CA, A1H0D |
 |
Soluble Glucose Dehydrogenase From Acinetobacter Calcoaceticus - Wild Type Ph8
Organism: Acinetobacter calcoaceticus
Method: X-RAY DIFFRACTION Resolution:1.19 Å Release Date: 2024-05-08 Classification: OXIDOREDUCTASE Ligands: A1H0D, CA, LI |
 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Homocysteine Bound.
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.39 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters And S-Adenosyl-L-Methionine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAM, EDO, GOL |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 5 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG, GOL |
 |
Crystal Structure Of Radical Sam Epimerase Epee C223A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Ripp Peptide 6 Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Crystal Structure Of Radical Sam Epimerase Epee D210A Mutant From Bacillus Subtilis With [4Fe-4S] Clusters, S-Adenosyl-L-Homocysteine And Persulfurated Cysteine Bound
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2024-01-10 Classification: METAL BINDING PROTEIN Ligands: SF4, SAH, EDO, PEG |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2022-04-13 Classification: HYDROLASE Ligands: EDO, NA |
 |
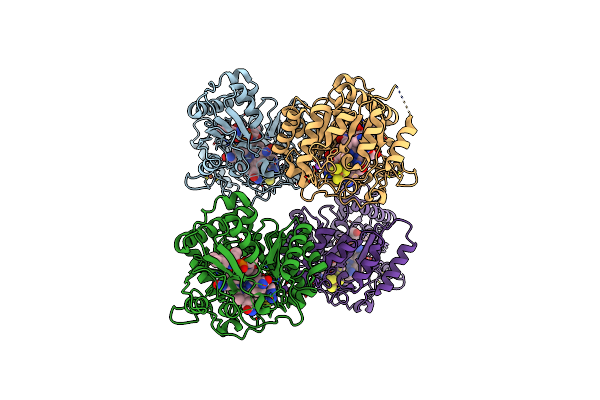
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, S-Adenosyl-L-Methionine, And Peptide Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, SAM, COB, NA |
 |
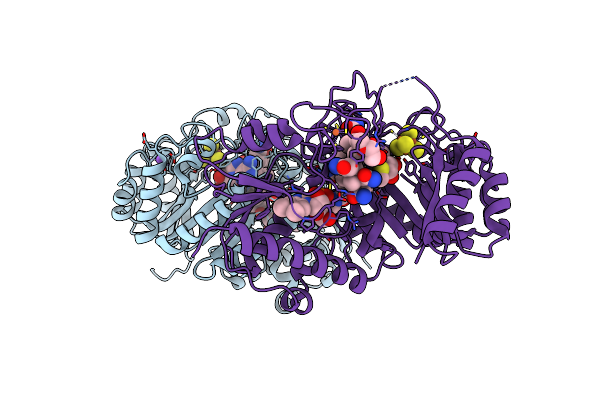
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Methyl-5'-Thioadenosine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, MTA, COB, NA |
 |
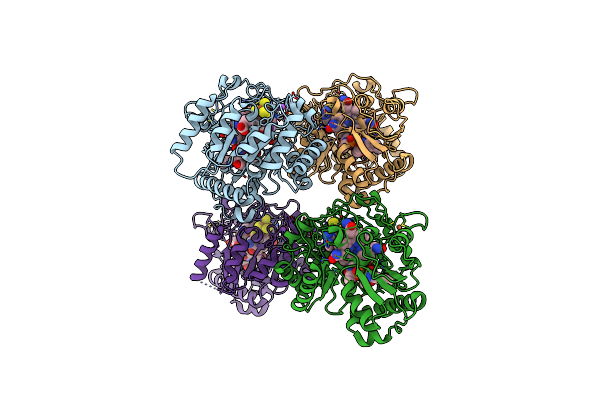
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Methyl-5'-Thioadenosine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, MTA, COB, NA, PEG |
 |
B12-Dependent Radical Sam Methyltransferase, Mmp10 With [4Fe-4S] Cluster, Cobalamin, And S-Adenosyl-L-Homocysteine Bound.
Organism: Methanosarcina acetivorans
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-02-02 Classification: METAL BINDING PROTEIN Ligands: SF4, FE, SAH, COB, NA |
 |
Crystal Structure Of The Dark-Adapted Full-Length Bacteriophytochrome Xccbphp-G454E Variant From Xanthomonas Campestris In The Pfr State
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2021-12-29 Classification: SIGNALING PROTEIN Ligands: BLA |
 |
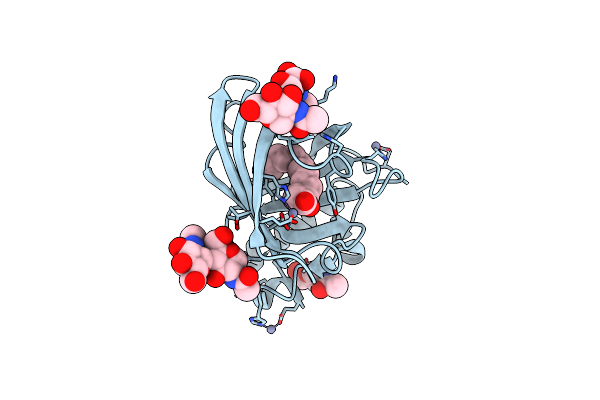
Crystal Structure Of The Photosensory Module From Xanthomonas Campestris Bacteriophytochrome Xccbphp In The Pfr State
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:2.95 Å Release Date: 2021-12-29 Classification: SIGNALING PROTEIN Ligands: BLA |
 |
Diploptera Punctata Inspired Lipocalin-Like Milk Protein Expressed In Saccharomyces Cerevisiae
Organism: Diploptera punctata
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2021-12-22 Classification: LIPID BINDING PROTEIN Ligands: NAG, PAM, GOL, ZN |
 |
Zn-Free Structure Of Lipocalin-Like Milk Protein, Inspired From Diploptera Punctata, Expressed In Saccharomyces Cerevisiae
Organism: Diploptera punctata
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2021-12-22 Classification: LIPID BINDING PROTEIN Ligands: PAM |
 |
Crystal Structure Of The Dark-Adapted Full-Length Bacteriophytochrome Xccbphp From Xanthomonas Campestris In The Pr State Bound To Bv Chromophore
Organism: Xanthomonas campestris pv. campestris (strain 8004)
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2020-12-30 Classification: SIGNALING PROTEIN Ligands: BLA |

