Search Count: 14
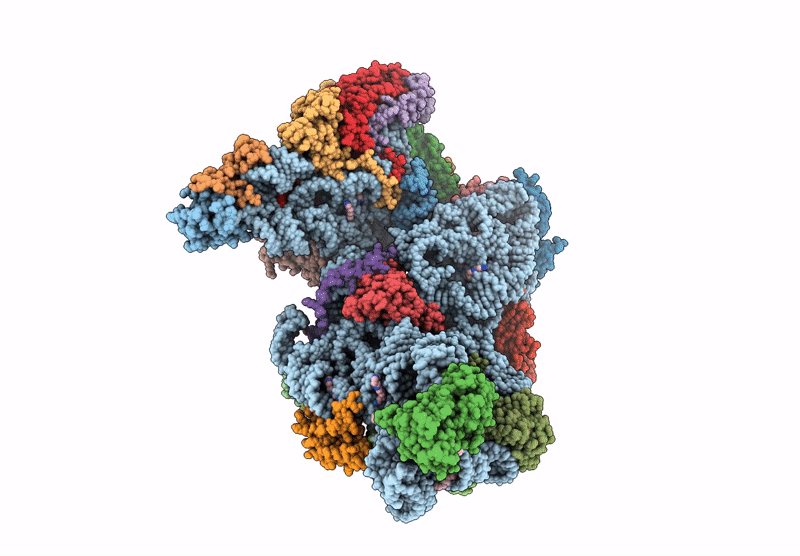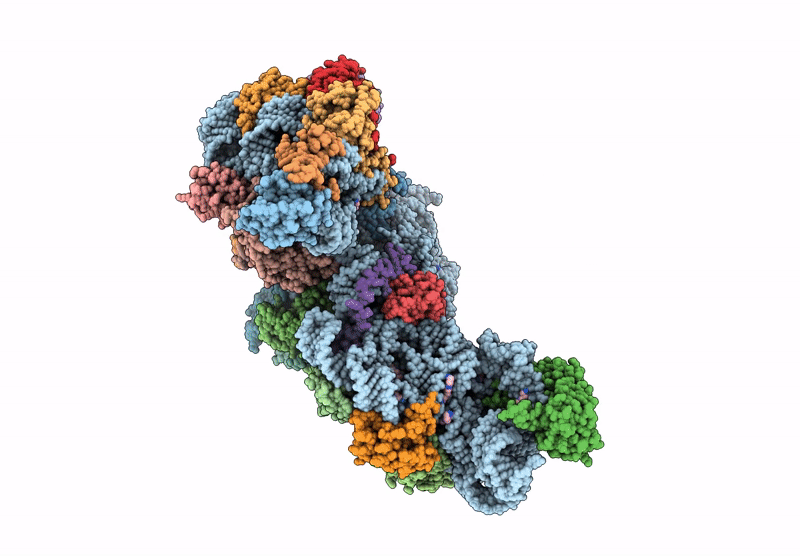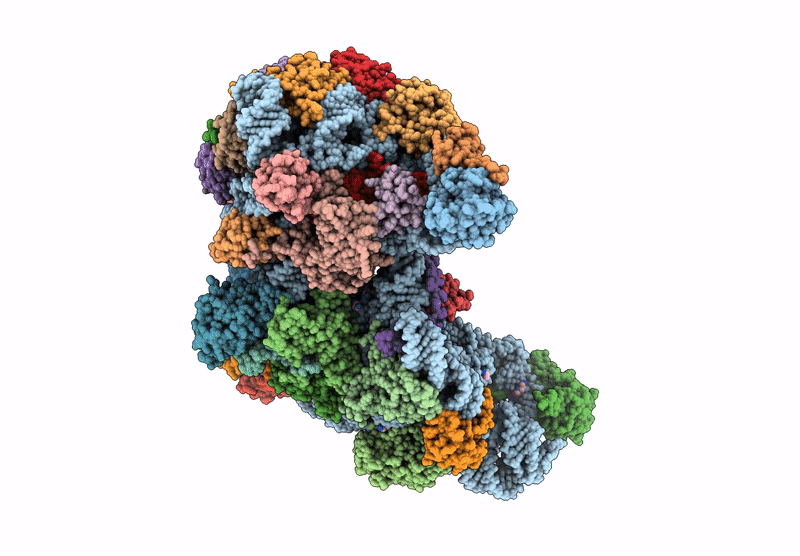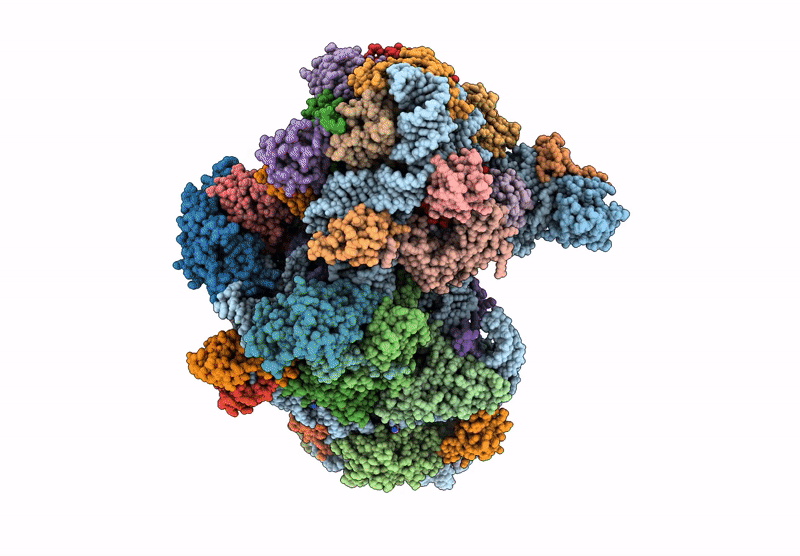 |
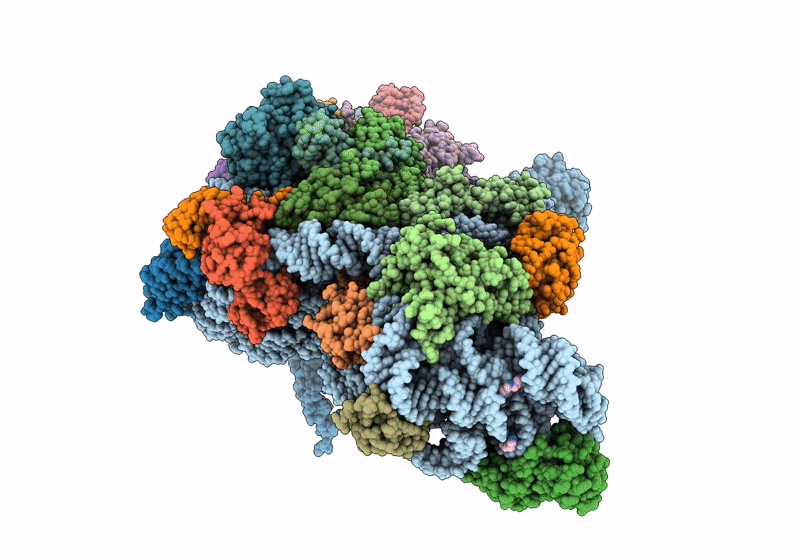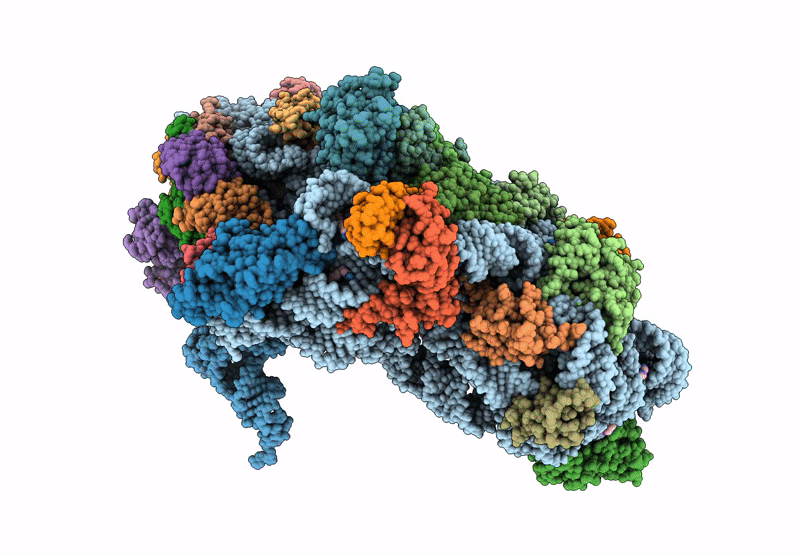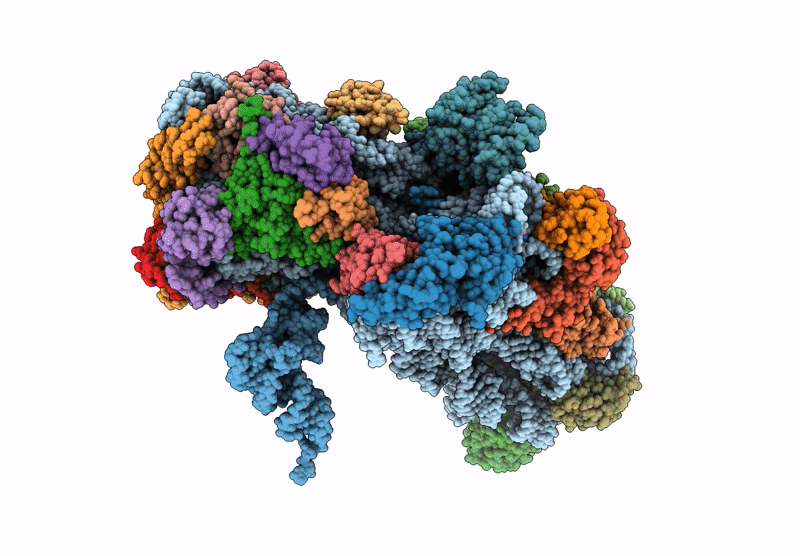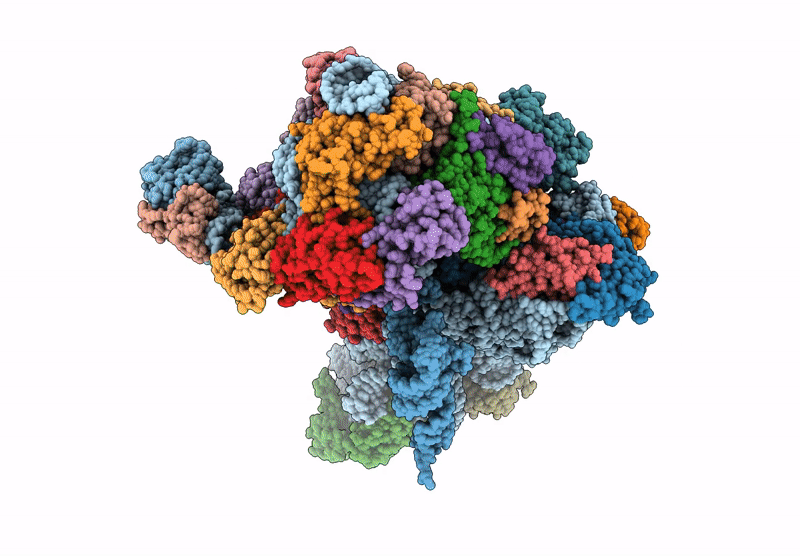
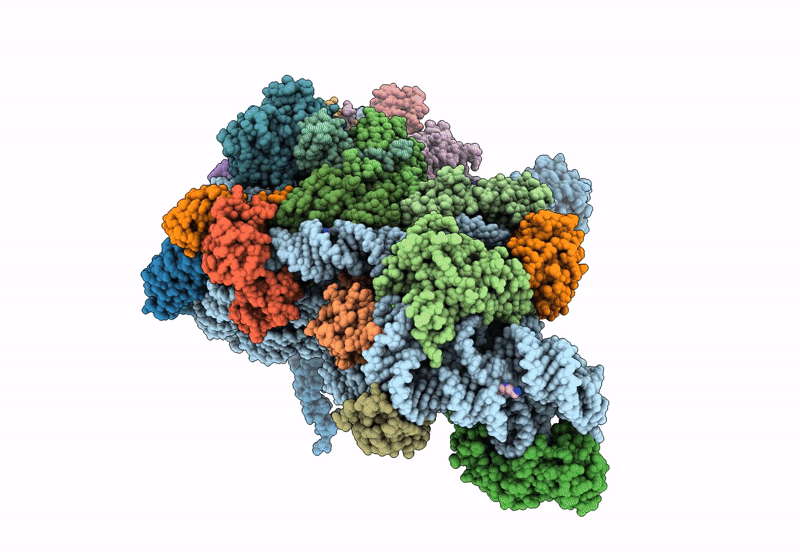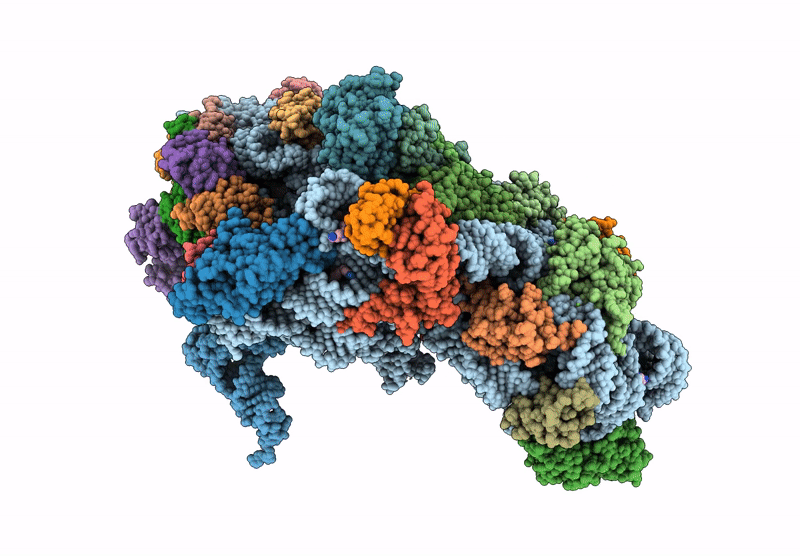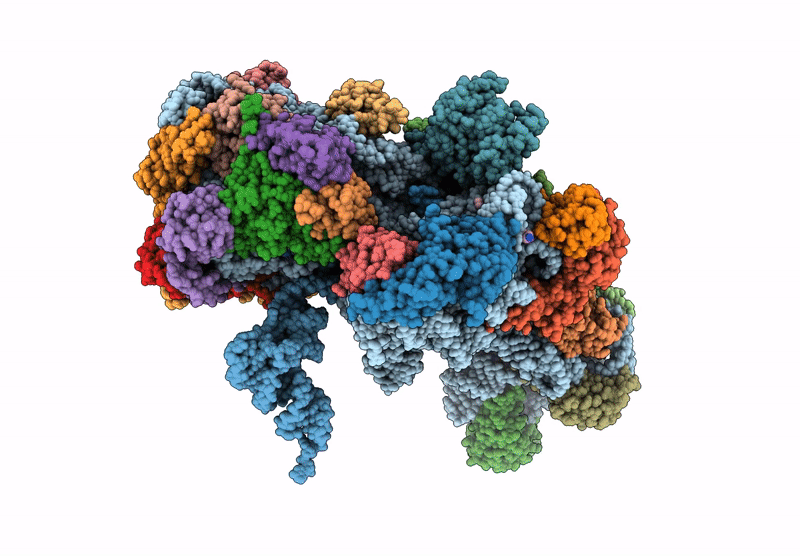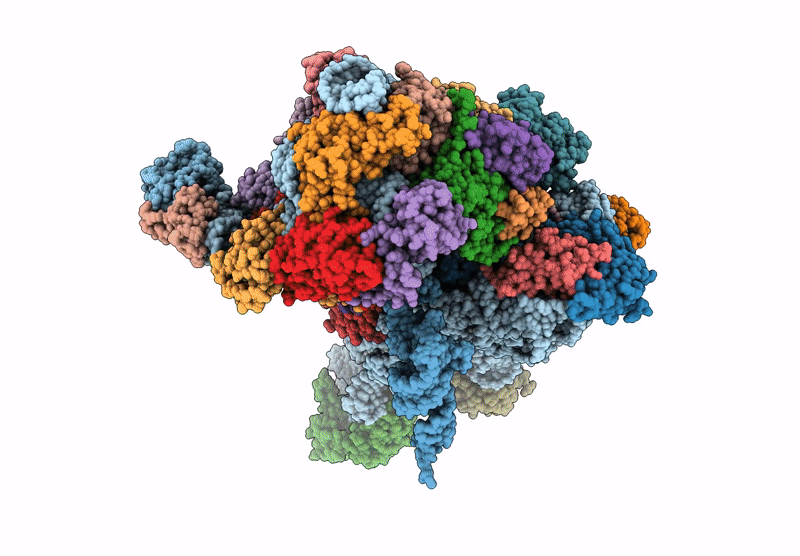
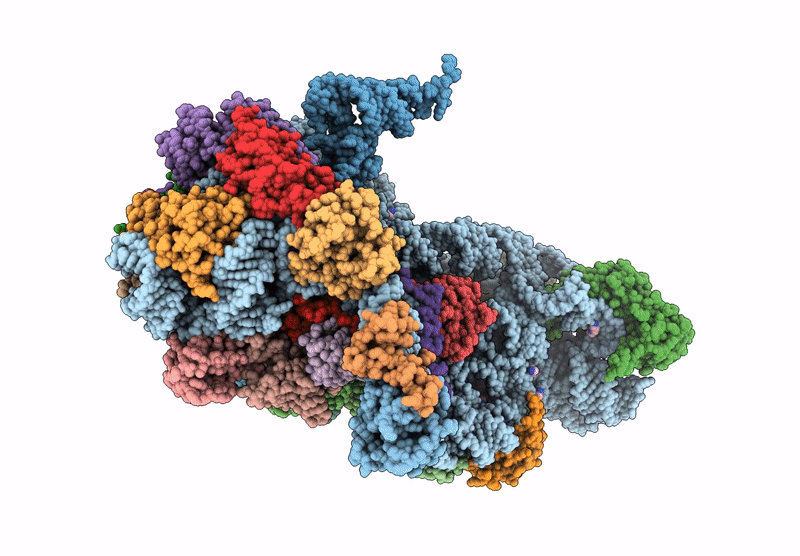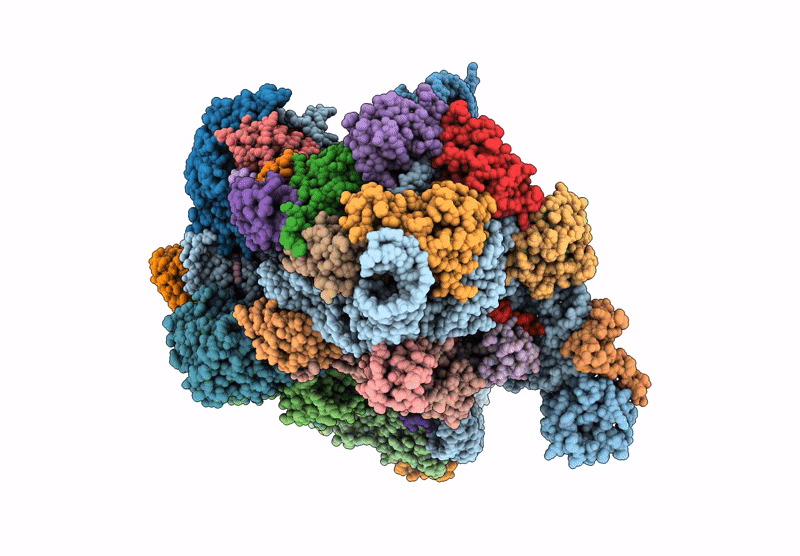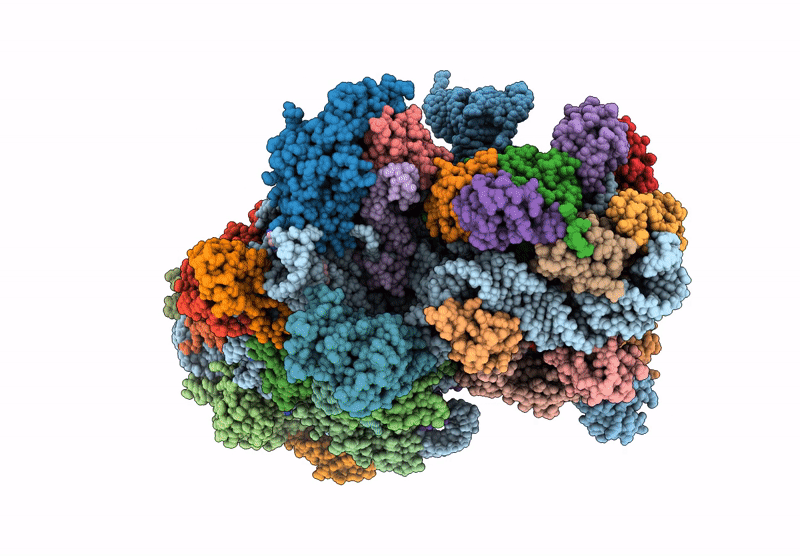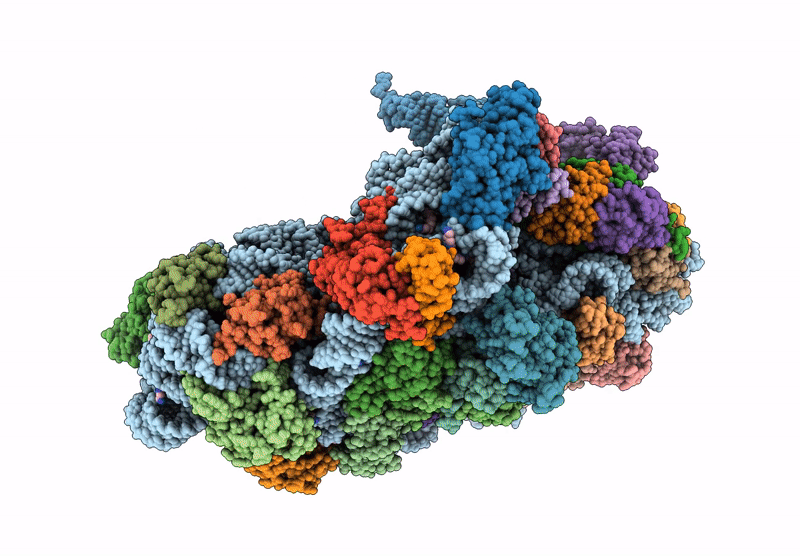
High-Resolution Cryo-Em Structure Of Saccharolobus Solfataricus 30S Ribosomal Subunit Bound To Mrna And Initiator Trna
Method: ELECTRON MICROSCOPY
Release Date: 2025-01-15 Classification: TRANSLATION Ligands: MG, SPM, ZN |
 |
Cryo-Em Structure Of Saccharolobus Solfataricus 30S Initiation Complex Bound To Ss-Map Leaderless Mrna
Organism: Escherichia coli, Saccharolobus solfataricus p2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION Ligands: MG, SPM, ZN |
 |
Cryo-Em Structure Of Saccharolobus Solfataricus 30S Initiation Complex Bound To Sd Mrna
Organism: Escherichia coli, Saccharolobus solfataricus p2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION Ligands: MG, SPM, ZN |
 |
Cryo-Em Structure Of Saccharolobus Solfataricus 30S Initiation Complex Bound To Sd Mrna With H44 In Up Position
Organism: Escherichia coli, Pyrococcus abyssi, Saccharolobus solfataricus p2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION Ligands: MG, SPM, ZN |
 |
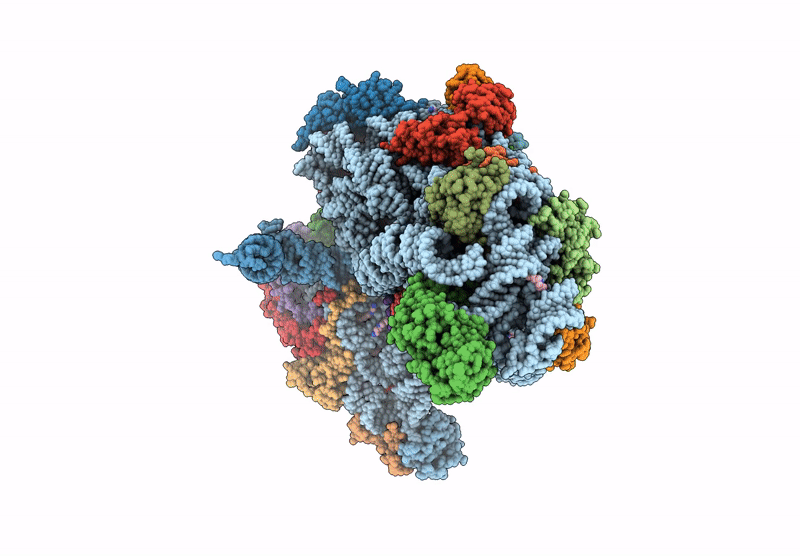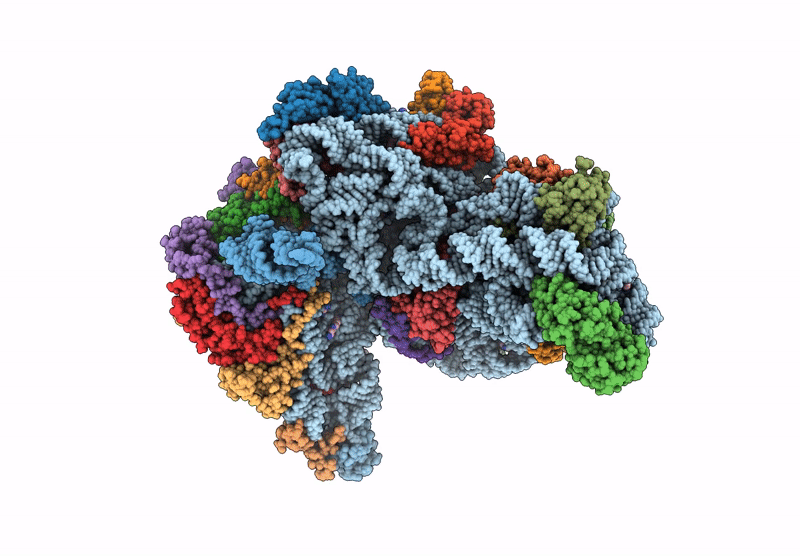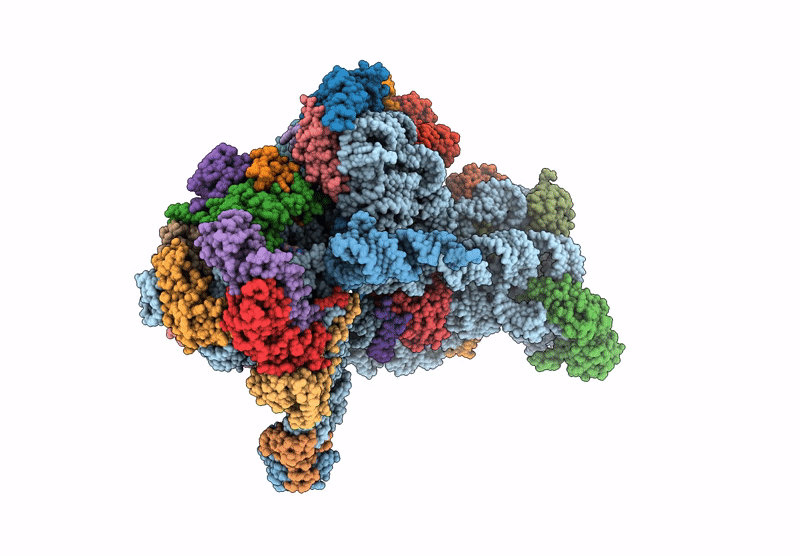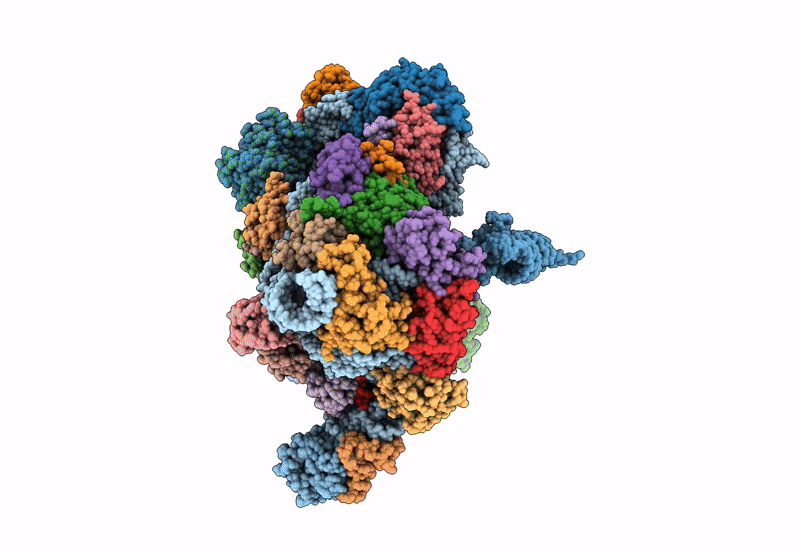
Cryo-Em Structure Of Saccharolobus Solfataricus 30S Initiation Complex Bound To Ss-Aif2Beta Leaderless Mrna With H44 In Up Position
Organism: Escherichia coli, Saccharolobus solfataricus p2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION Ligands: SPM, MG, ZN, GTP |
 |
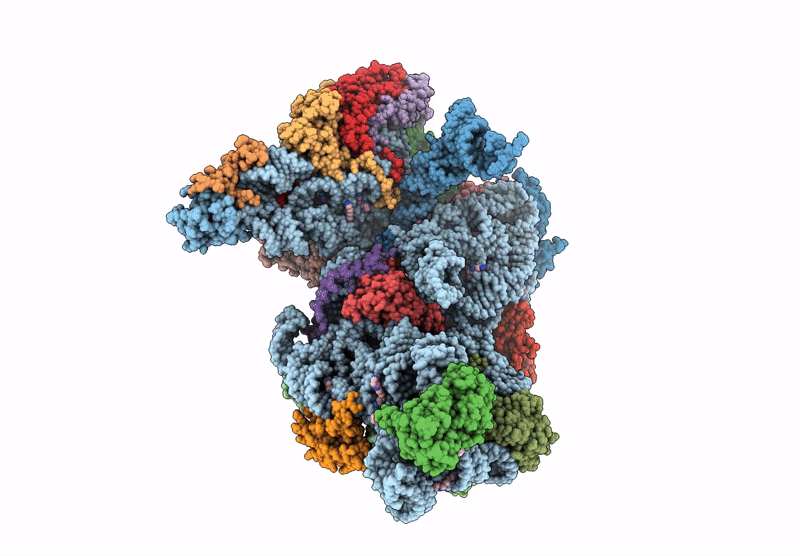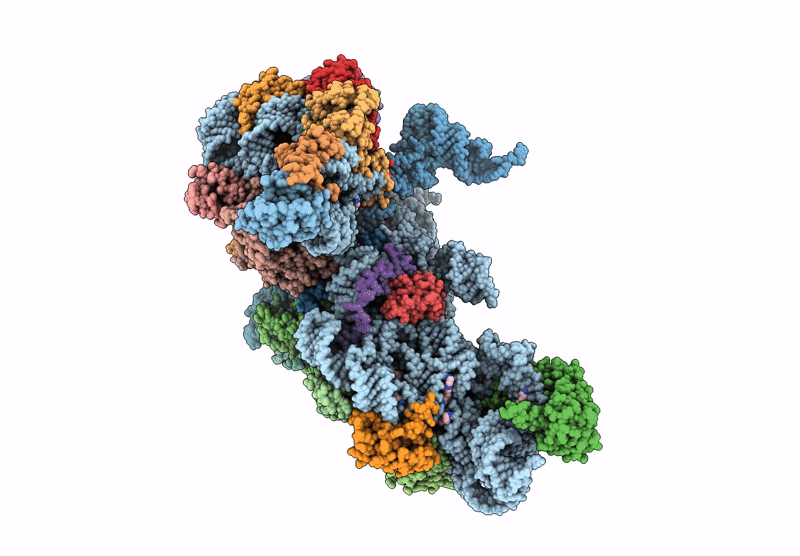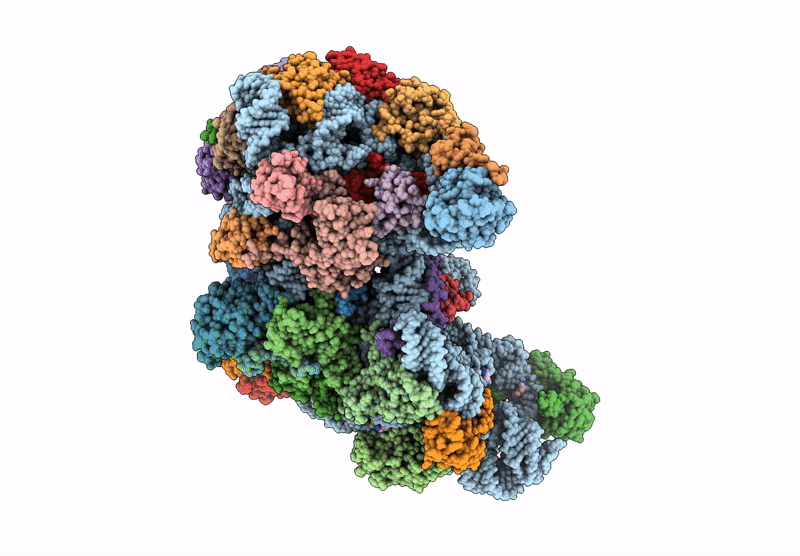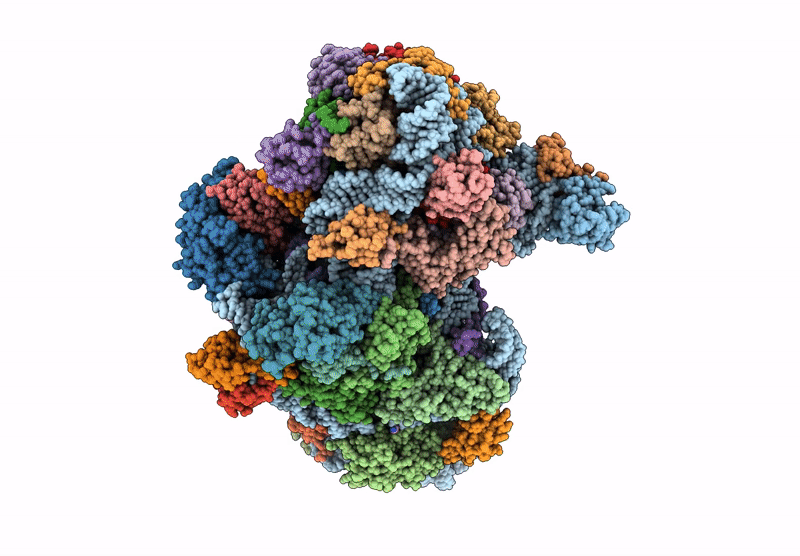
Cryo-Em Structure Of Saccharolobus Solfataricus 30S Initiation Complex Bound To Ss-Aef1A-Like Mrna
Organism: Escherichia coli, Saccharolobus solfataricus p2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION Ligands: MG, SPM, ZN |
 |
Cryo-Em Structure Of Saccharolobus Solfataricus 30S Initiation Complex Bound To Ss-Map Leaderless Mrna With H44 In Up Position
Organism: Escherichia coli, Saccharolobus solfataricus p2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION Ligands: SPM, MG, ZN, ATP |
 |
Cryo-Em Structure Of Saccharolobus Solfataricus 30S Initiation Complex Bound To Ss-Aif2Beta Leaderless Mrna
Organism: Escherichia coli, Saccharolobus solfataricus p2
Method: ELECTRON MICROSCOPY Release Date: 2025-01-15 Classification: TRANSLATION Ligands: SPM, MG, ZN, GTP |
 |
Crystal Structure Of The Ptpn3 Pdz Domain Bound To The Hbv Core Protein C-Terminal Peptide
Organism: Homo sapiens, Hepatitis b virus
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2021-01-20 Classification: HYDROLASE Ligands: BR |
 |
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2020-04-22 Classification: HYDROLASE Ligands: DIO |
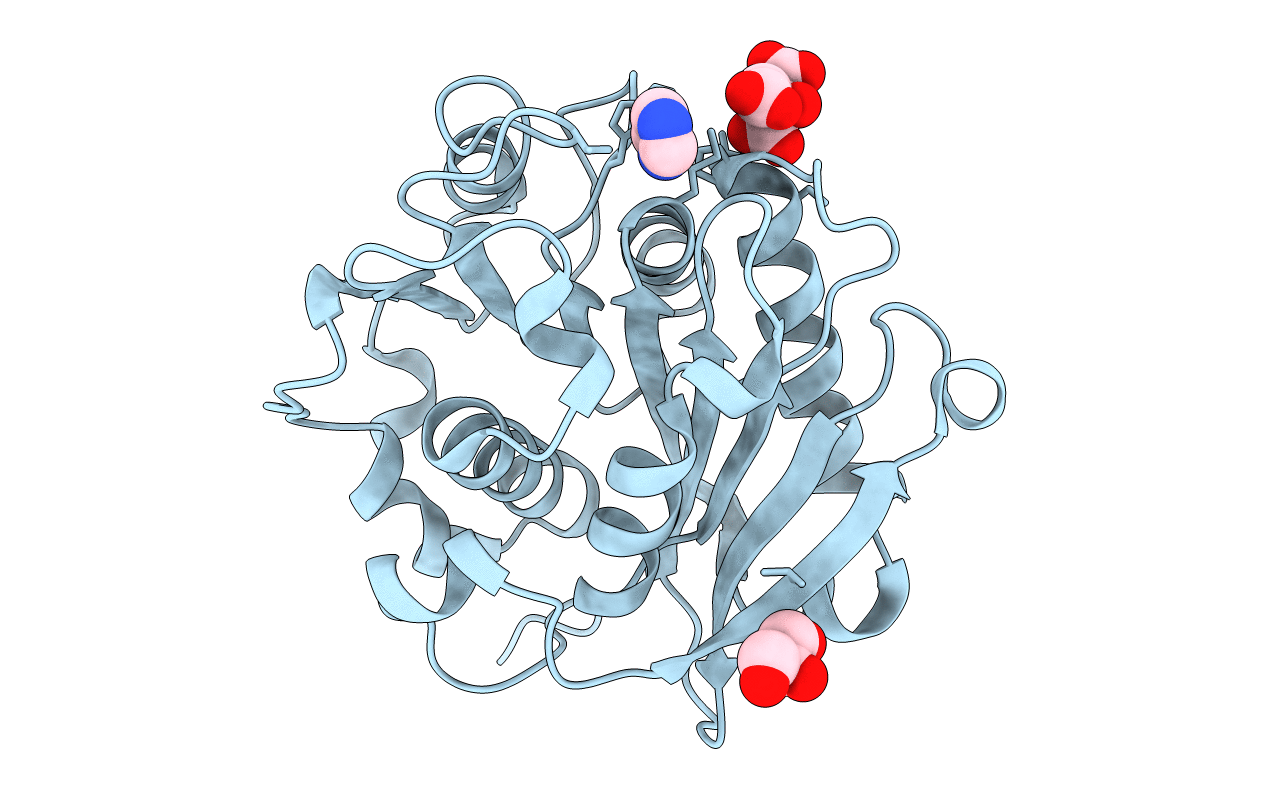 |
High Resolution Crystal Structure Of A Leaf-Branch Compost Cutinase Quintuple Variant
Organism: Uncultured bacterium
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2020-04-15 Classification: HYDROLASE Ligands: IMD, GOL, CIT |
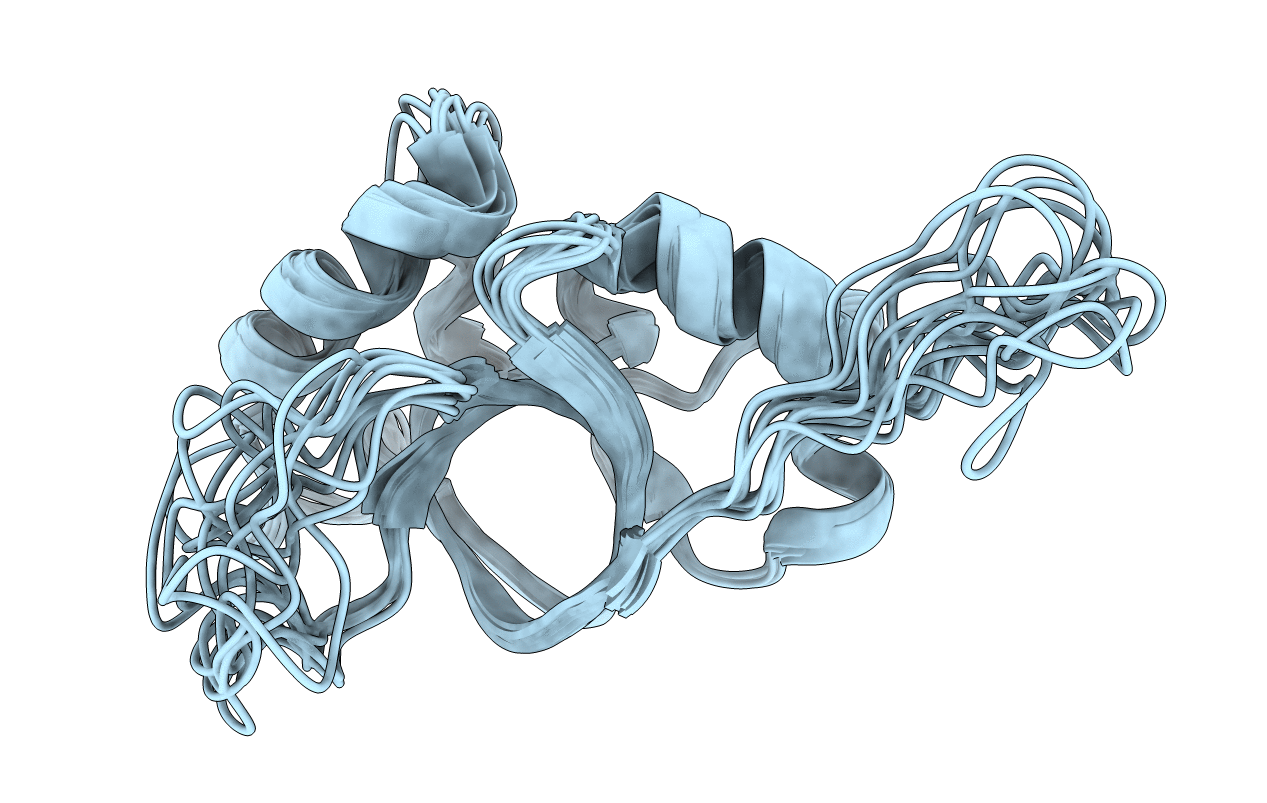 |
Organism: Neosartorya fumigata (strain cea10 / cbs 144.89 / fgsc a1163)
Method: SOLUTION NMR Release Date: 2019-03-27 Classification: STRUCTURAL PROTEIN |
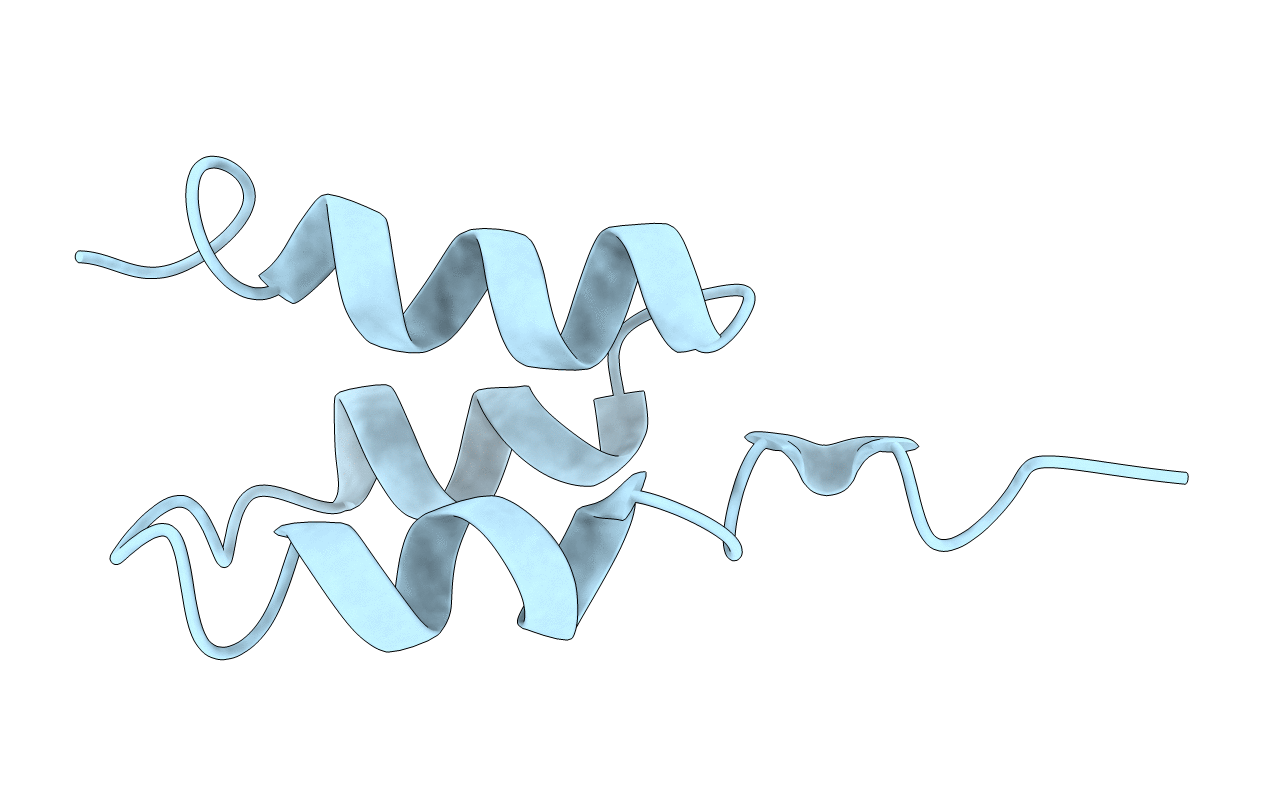 |
Organism: Finegoldia magna atcc 29328
Method: SOLUTION NMR Release Date: 1997-07-23 Classification: ALBUMIN-BINDING PROTEIN |
 |
Organism: Finegoldia magna atcc 29328
Method: SOLUTION NMR Release Date: 1997-07-07 Classification: ALBUMIN-BINDING PROTEIN |

