Search Count: 17
 |
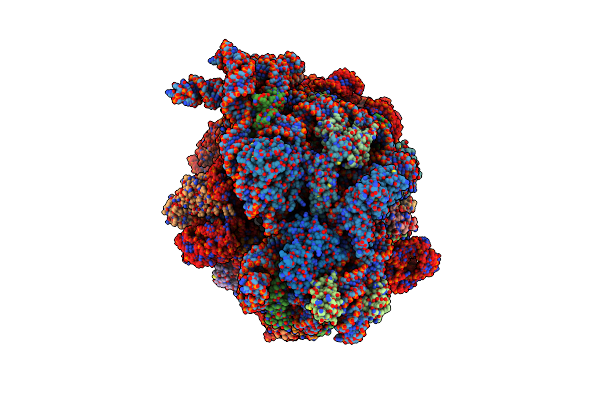
Organism: Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: RIBOSOME Ligands: MG, ZN |
 |
Structure Of The Higb1 Toxin Mutant K95A From Mycobacterium Tuberculosis (Rv1955) And Its Target, The Cspa Mrna, On The E. Coli Ribosome.
Organism: Mycobacterium tuberculosis h37rv, Escherichia coli, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2022-03-02 Classification: TOXIN Ligands: MG, FME, ZN |
 |
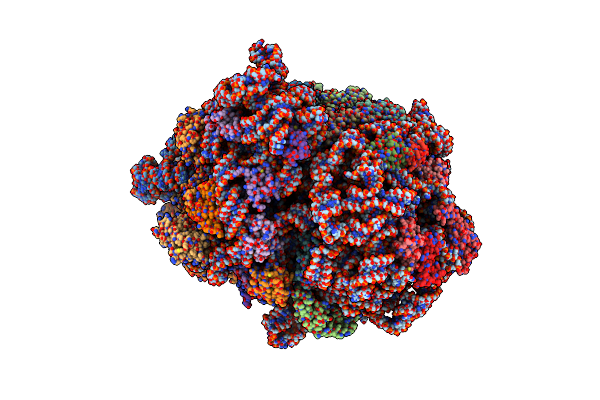
Crystal Structure Of The Higb1 Toxin Mutant K95A From Mycobacterium Tuberculosis (Rv1955)
Organism: Mycobacterium tuberculosis (strain atcc 25618 / h37rv)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-11-24 Classification: TOXIN |
 |
Structure Of Pre-Accomodated Trans-Translation Complex On E. Coli Stalled Ribosome.
Method: ELECTRON MICROSCOPY
Release Date: 2021-08-18 Classification: TRANSLATION Ligands: MG, KIR, GDP, ZN |
 |
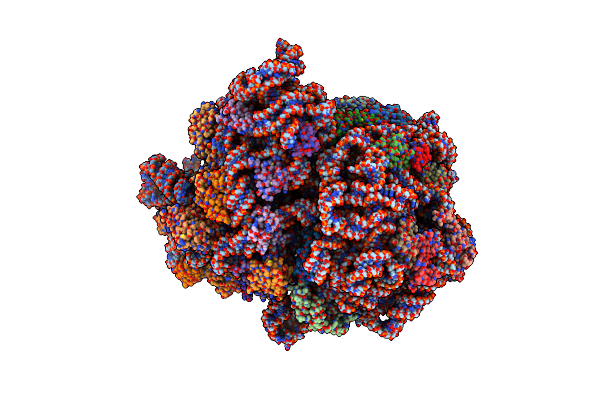
Structure Of Accomodated Trans-Translation Complex On E. Coli Stalled Ribosome.
Organism: Escherichia coli, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: TRANSLATION Ligands: MG, ZN |
 |
Structure Of Translocated Trans-Translation Complex On E. Coli Stalled Ribosome.
Organism: Escherichia coli bl21(de3), Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: TRANSLATION Ligands: MG, ZN |
 |
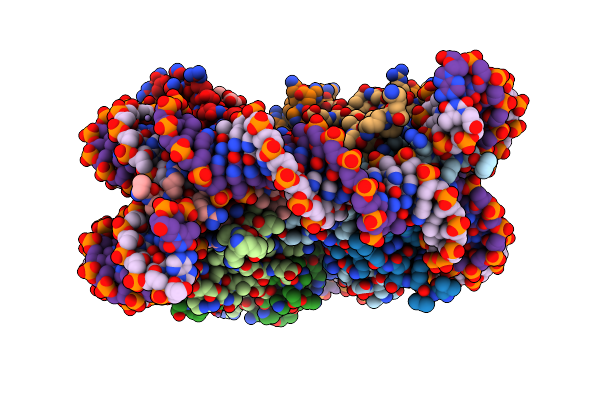
Structure Of Post-Translocated Trans-Translation Complex On E. Coli Stalled Ribosome.
Organism: Escherichia coli, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-08-18 Classification: TRANSLATION Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-01-15 Classification: DNA BINDING PROTEIN/DNA Ligands: CL, MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-01-15 Classification: DNA BINDING PROTEIN/DNA Ligands: MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2020-01-15 Classification: DNA BINDING PROTEIN/DNA Ligands: CL, MN, K |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.85 Å Release Date: 2020-01-15 Classification: DNA BINDING PROTEIN/DNA Ligands: CL, MN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-01-15 Classification: DNA BINDING PROTEIN/DNA Ligands: CL, MN, K |
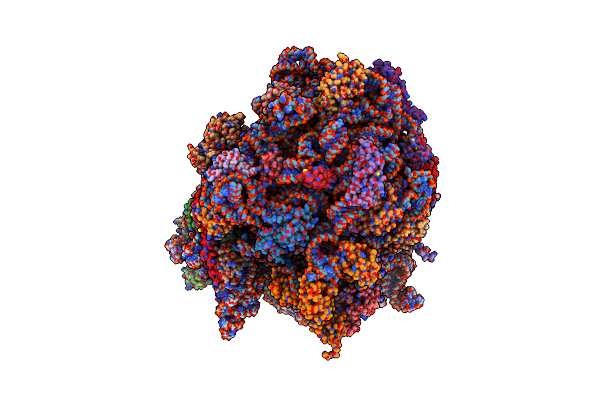 |
Elongation Factor G-Ribosome Complex Captures In The Absence Of Inhibitors.
Organism: Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579), Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2018-02-14 Classification: RIBOSOME Ligands: MG, ZN, GDP |
 |
Organism: Bacillus thuringiensis bt407
Method: X-RAY DIFFRACTION Resolution:3.24 Å Release Date: 2016-07-13 Classification: SIGNALING PROTEIN |
 |
Organism: Bacillus thuringiensis serovar thuringiensis
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2013-07-03 Classification: Transcription, Peptide binding protein |
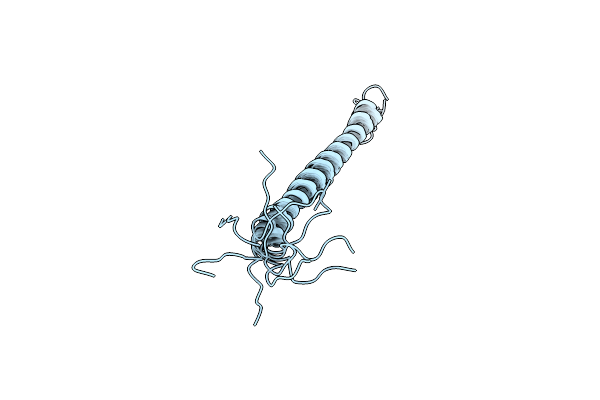 |
Association Of Subunit D (Vma6P) And E (Vma4P) With G (Vma10P) And The Nmr Solution Structure Of Subunit G (G1-59) Of The Saccharomyces Cerevisiae V1Vo Atpase
Organism: Saccharomyces cerevisiae
Method: SOLUTION NMR Release Date: 2009-08-11 Classification: HYDROLASE |
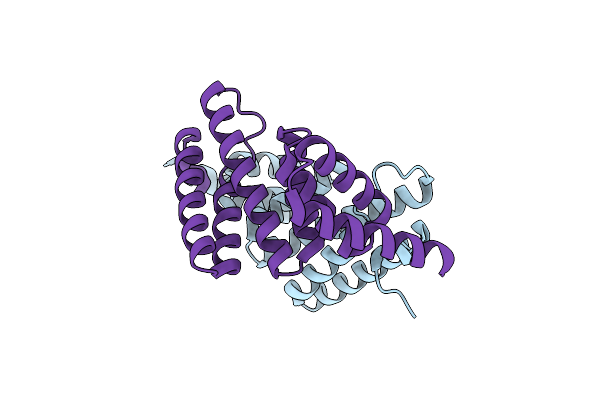 |
Plasmodium Falciparum Putative Fk506-Binding Protein Pfl2275C, C-Terminal Tpr-Containing Domain
Organism: Plasmodium falciparum
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2006-01-24 Classification: STRUCTURAL GENOMICS, UNKNOWN FUNCTION |

