Search Count: 25
 |
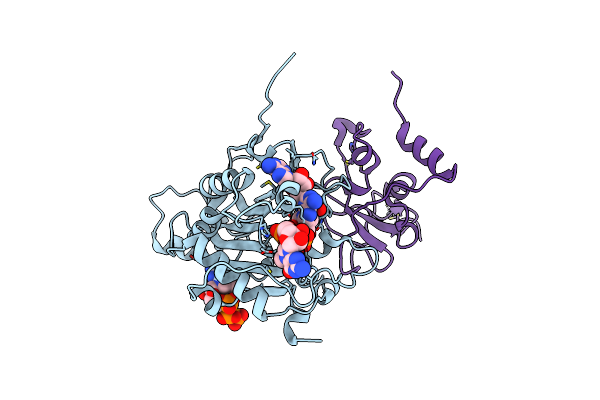
Room-Temperature X-Ray Structure Of Sars-Cov-2 Main Protease (3Cl Mpro) In Complex With A Non-Covalent Inhibitor Mcule-5948770040
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-03-03 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: YD1 |
 |
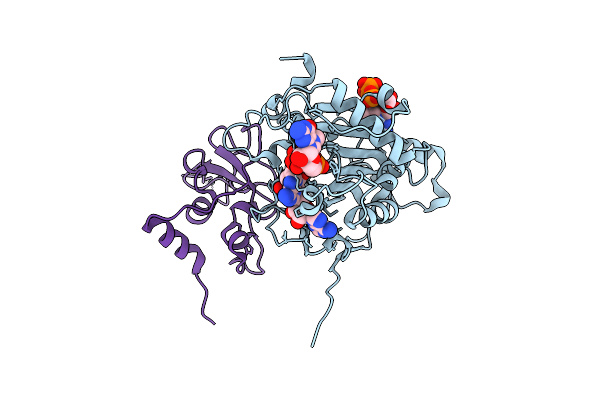
Room Temperature Structure Of Sars-Cov-2 Nsp10/Nsp16 Methyltransferase In A Complex With 2'-O-Methylated M7Gpppa Cap-1 And Sah Determined By Fixed-Target Serial Crystallography
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-08-26 Classification: VIRAL PROTEIN Ligands: CL, SAH, V9G, MGP, ZN |
 |
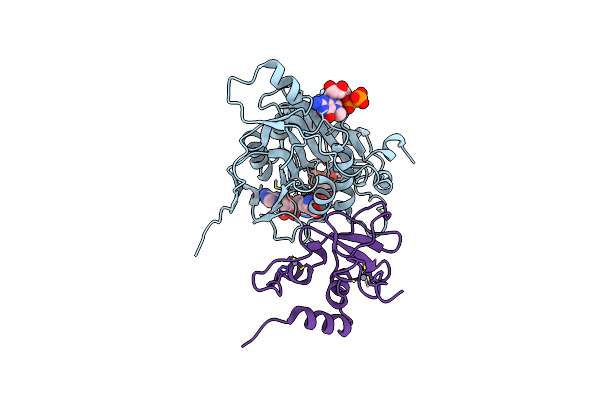
Room Temperature Crystal Structure Of Nsp10/Nsp16 From Sars-Cov-2 With Substrates And Products Of 2'-O-Methylation Of The Cap-1
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.65 Å Release Date: 2020-08-26 Classification: VIRAL PROTEIN Ligands: CL, SAM, SAH, GTA, V9G, MGP, ZN |
 |
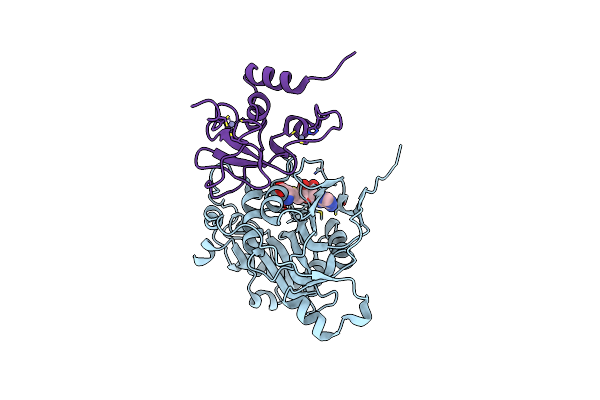
Room Temperature Structure Of Sars-Cov-2 Nsp10/Nsp16 Methyltransferase In A Complex With M7Gpppa Cap-0 And Sam Determined By Fixed-Target Serial Crystallography
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.18 Å Release Date: 2020-08-26 Classification: VIRAL PROTEIN Ligands: GTA, SAM, M7G, ZN |
 |
Room Temperature Structure Of Sars-Cov-2 Nsp10/Nsp16 Methyltransferase In A Complex With Sam Determined By Fixed-Target Serial Crystallography
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-07-08 Classification: VIRAL PROTEIN Ligands: CL, SAM, ZN |
 |
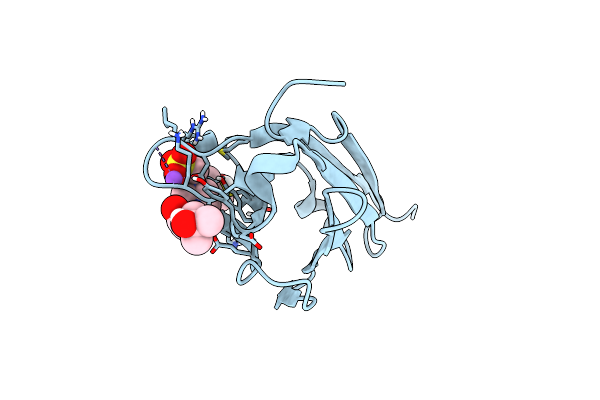
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN/LIPID BINDING PROTEIN Ligands: EDO, CL, MG |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2018-09-12 Classification: LIPID BINDING PROTEIN Ligands: PC, CA |
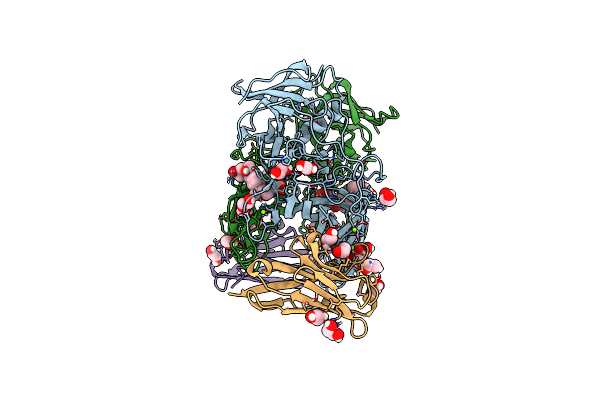 |
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor And Glycochenodeoxycholic Acid
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN Ligands: MG, EDO, CHO |
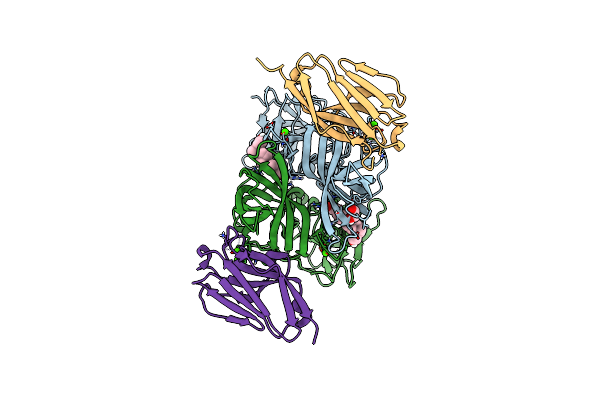 |
Crystal Structure Of The Murine Norovirus Vp1 P Domain In Complex With The Cd300Lf Receptor And Lithocholic Acid
Organism: Murine norovirus 1, Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-09-12 Classification: VIRAL PROTEIN Ligands: 4OA, CA, EDO |
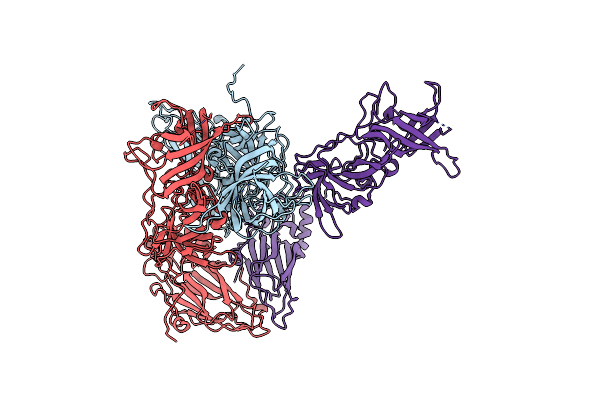 |
Mouse Norovirus Model Using The Crystal Structure Of Mnv P Domain And The Norwalkvirus Shell Domain
Organism: Norwalk virus, Murine norovirus 1
Method: ELECTRON MICROSCOPY Release Date: 2018-04-04 Classification: VIRUS |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2016-08-31 Classification: IMMUNE SYSTEM Ligands: EPE, NA, MPD |
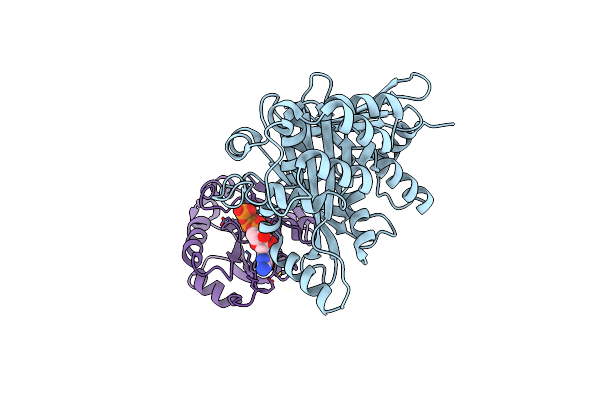 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-01-05 Classification: PROTEIN TRANSPORT Ligands: GTP, MG |
 |
Organism: Escherichia coli, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.86 Å Release Date: 2011-01-05 Classification: PROTEIN TRANSPORT/TRANSFERASE |
 |
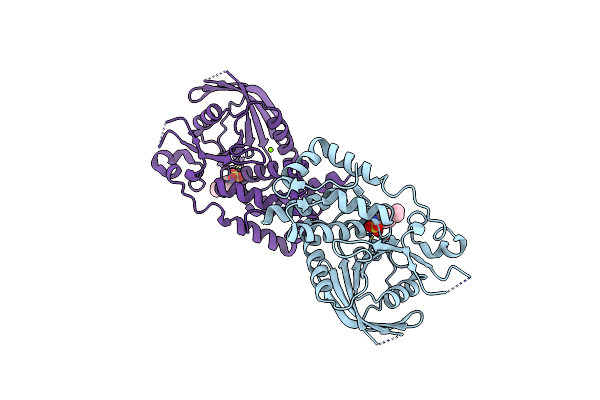
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-07-21 Classification: SIGNALING PROTEIN/TRANSCRIPTION |
 |
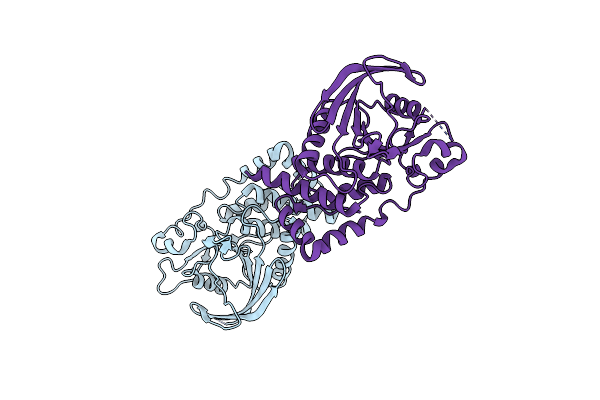
Engineering The Ptpbeta Catalytic Domain With Improved Crystallization Properties
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2006-09-05 Classification: HYDROLASE Ligands: CL, UA5, MG |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-08-29 Classification: HYDROLASE Ligands: CL |
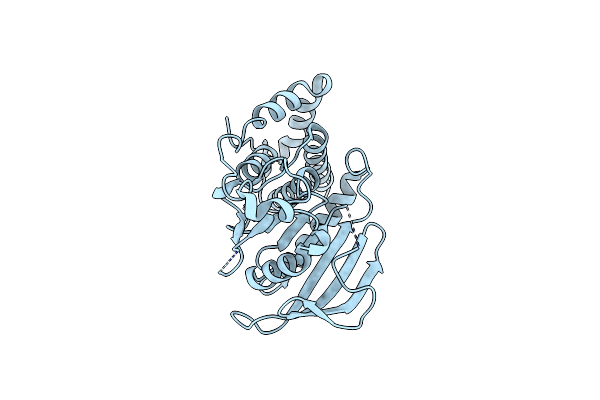 |
Structural Studies Of Protein Tyrosine Phosphatase Beta Catalytic Domain In Complex With Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2006-08-29 Classification: HYDROLASE |
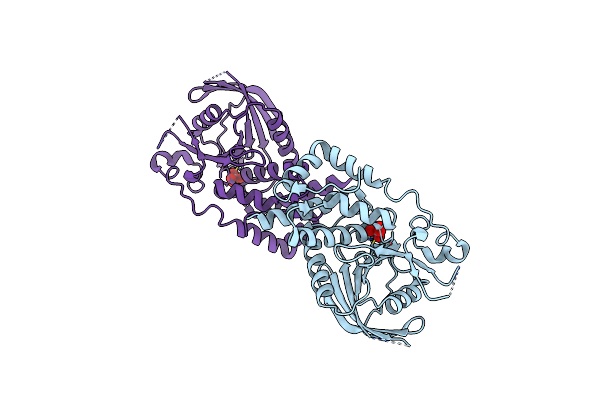 |
Structural Studies Of Protein Tyrosine Phosphatase Beta Catalytic Domain In Complex With Inhibitors
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2006-08-29 Classification: HYDROLASE Ligands: VO4 |
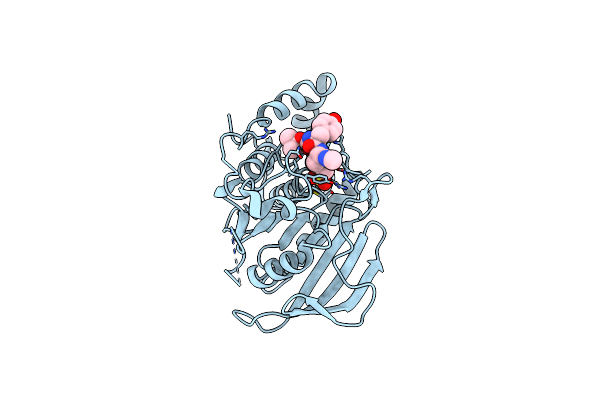 |
Structural Studies Of Protein Tyrosine Phosphatase Beta Catalytic Domain In Complex With A Sulfamic Acid (Soaking Experiment)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2006-08-29 Classification: HYDROLASE Ligands: CL, UA1 |
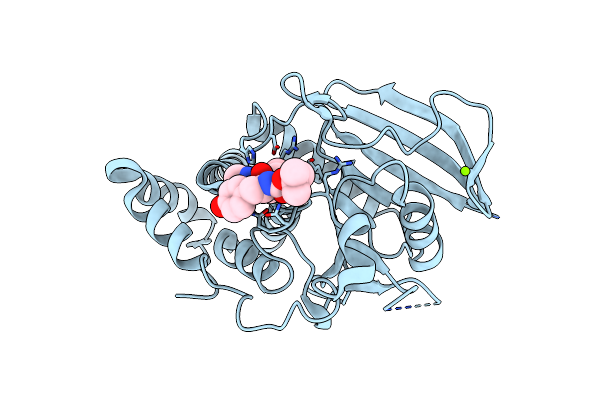 |
Structural Studies Of Protein Tyrosine Phosphatase Beta Catalytic Domain Co-Crystallized With A Sulfamic Acid Inhibitor
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2006-08-29 Classification: HYDROLASE Ligands: MG, CL, UA1 |

