Search Count: 52
 |
Organism: Adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2022-11-23 Classification: VIRUS LIKE PARTICLE |
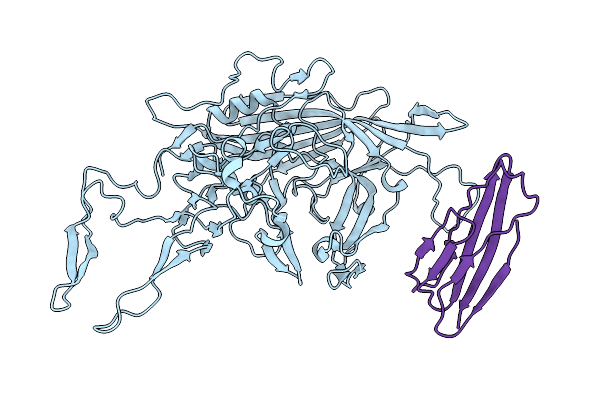 |
Adeno-Associated Virus Go.1 In Complex With Its Cellular Receptor Aavr At 2.4 Angstroms Resolution, Aavgo.1 Aavr
Organism: Homo sapiens, Adeno-associated virus
Method: ELECTRON MICROSCOPY Resolution:2.40 Å Release Date: 2022-11-23 Classification: VIRUS LIKE PARTICLE |
 |
Adeno-Associated Virus (Aav-Dj) - Cryo-Em Structure At 1.56 Angstrom Resolution
Organism: Adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2020-12-16 Classification: VIRUS Ligands: MG |
 |
Organism: Adeno-associated virus - 5
Method: ELECTRON MICROSCOPY Resolution:2.10 Å Release Date: 2020-12-16 Classification: VIRUS |
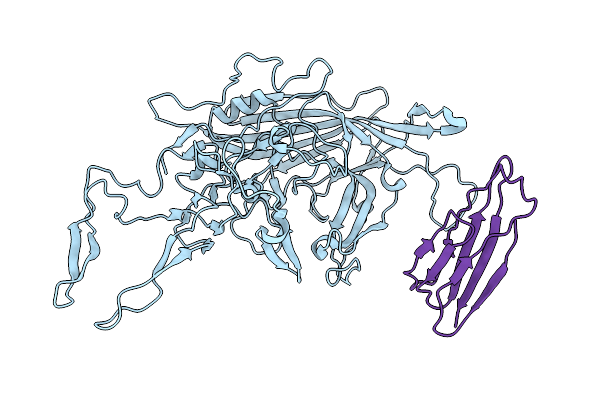 |
Adeno-Associated Virus Serotype 5 In Complex With The Cellular Receptor Aavr At 2.5 Angstroms Resolution, Aav5 Aavr
Organism: Homo sapiens, Adeno-associated virus - 5
Method: ELECTRON MICROSCOPY Resolution:2.50 Å Release Date: 2020-12-16 Classification: VIRUS |
 |
Organism: Homo sapiens, Adeno-associated virus - 2
Method: ELECTRON MICROSCOPY Release Date: 2019-06-12 Classification: VIRUS Ligands: MG |
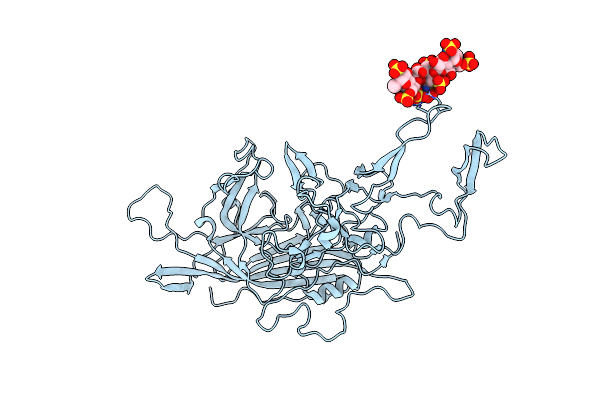 |
The 2.8 A Electron Microscopy Structure Of Adeno-Associated Virus-Dj Bound By A Heparanoid Pentasaccharide
Organism: Adeno-associated virus
Method: ELECTRON MICROSCOPY Release Date: 2017-05-24 Classification: VIRUS LIKE PARTICLE |
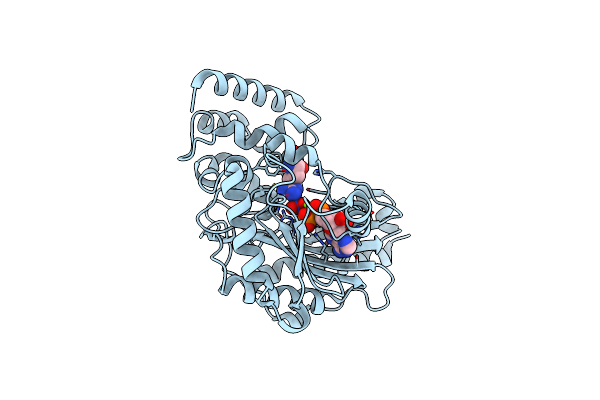 |
Ambient Temperature Transition State Structure Of Arginine Kinase - Crystal 8/Form I
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2016-08-17 Classification: TRANSFERASE Ligands: ADP, ARG, NO3, MG |
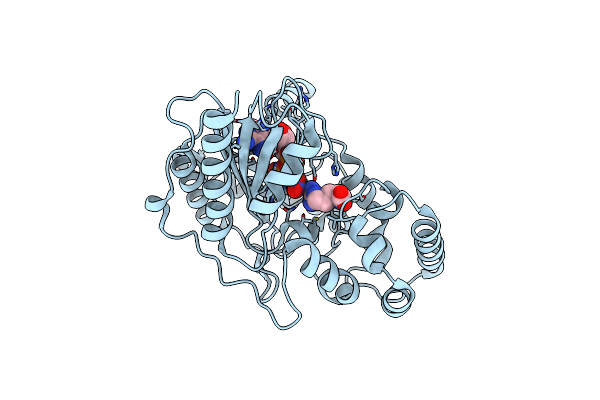 |
Ambient Temperature Transition State Structure Of Arginine Kinase - Crystal 11/Form Ii
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-08-17 Classification: TRANSFERASE Ligands: NO3, MG, ARG, ADP |
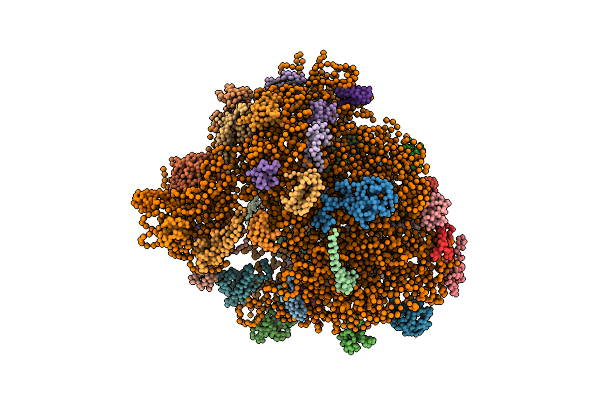 |
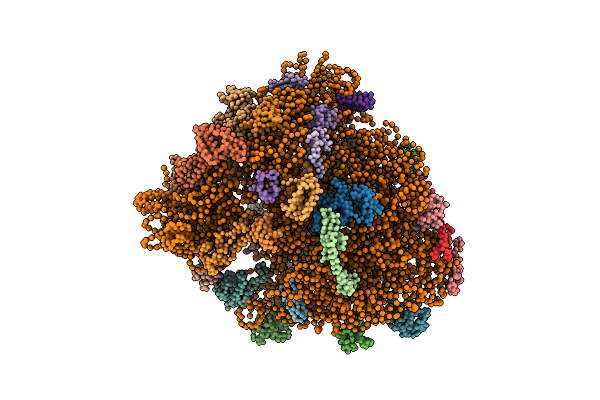
Real Space Refined Coordinates Of The 30S And 50S Subunits Fitted Into The Low Resolution Cryo-Em Map Of The Ef-G.Gtp State Of E. Coli 70S Ribosome
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
 |
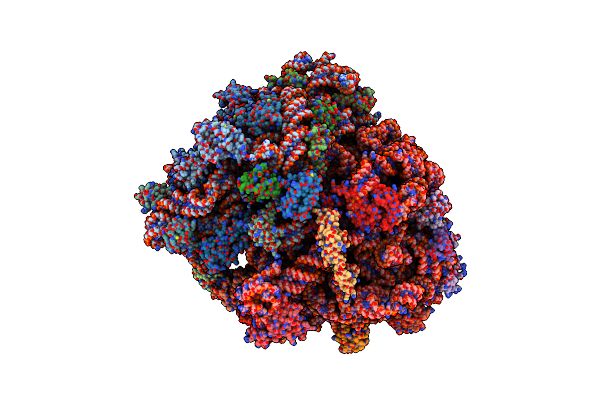
Real Space Refined Coordinates Of The 30S And 50S Subunits Fitted Into The Low Resolution Cryo-Em Map Of The Initiation-Like State Of E. Coli 70S Ribosome
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
 |
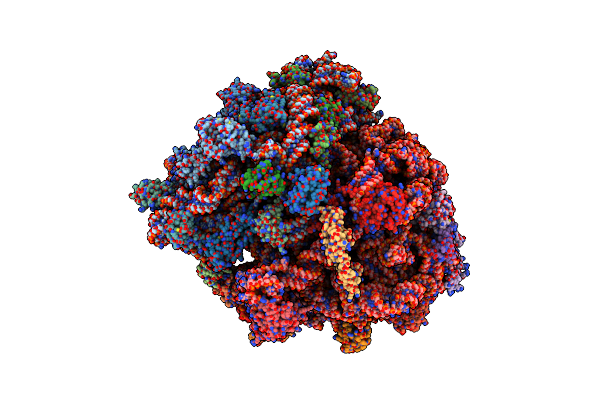
Structure Of A Pre-Translocational E. Coli Ribosome Obtained By Fitting Atomic Models For Rna And Protein Components Into Cryo-Em Map Emd-1056
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
 |
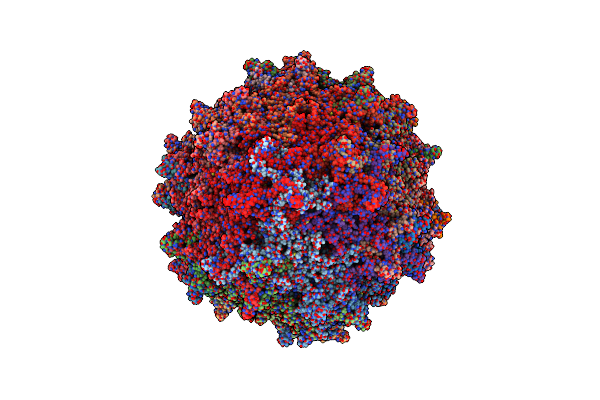
Structure Of A Secm-Stalled E. Coli Ribosome Complex Obtained By Fitting Atomic Models For Rna And Protein Components Into Cryo-Em Map Emd-1143
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2014-07-09 Classification: RIBOSOME |
 |
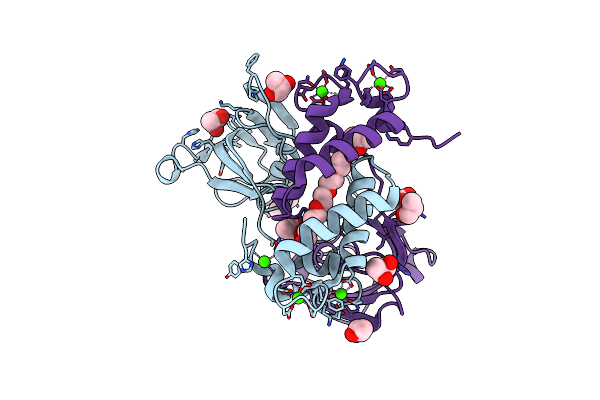
Structure-Function Analysis Of Receptor-Binding In Adeno-Associated Virus Serotype 6 (Aav-6)
Organism: Adeno-associated virus - 6
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2014-07-09 Classification: VIRUS |
 |
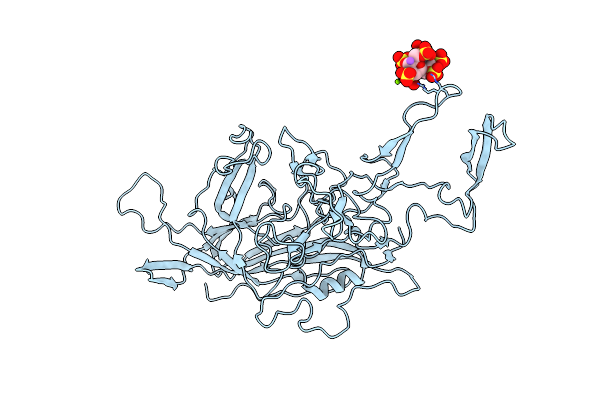
Crystal Structure Of Human Peptidyl-Prolyl Cis-Trans Isomerase Fkbp22 (Aka Fkbp14) Containing Two Ef-Hand Motifs
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2013-12-25 Classification: ISOMERASE Ligands: CA, 1PE, PGO |
 |
Electron Microscopy Analysis Of A Disaccharide Analog Complex Reveals Receptor Interactions Of Adeno-Associated Virus
Organism: Adeno-associated virus
Method: ELECTRON MICROSCOPY Resolution:4.80 Å Release Date: 2013-10-16 Classification: VIRUS |
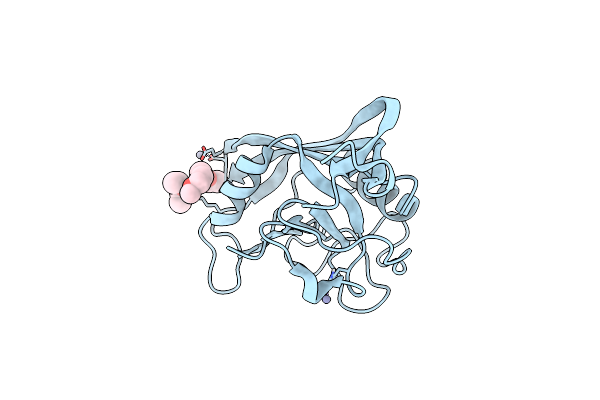 |
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-11-14 Classification: ISOMERASE Ligands: ZN, ME2, PEG |
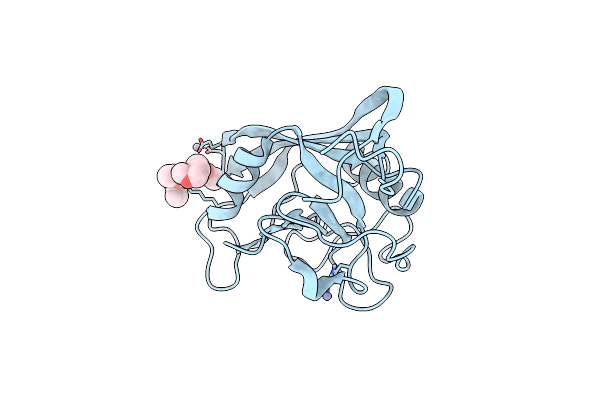 |
Crystal Structure Of Mutated Cyclophilin B That Causes Hyperelastosis Cutis In The American Quarter Horse
Organism: Equus caballus
Method: X-RAY DIFFRACTION Resolution:1.10 Å Release Date: 2012-11-14 Classification: ISOMERASE Ligands: ZN, ME2, PEG |
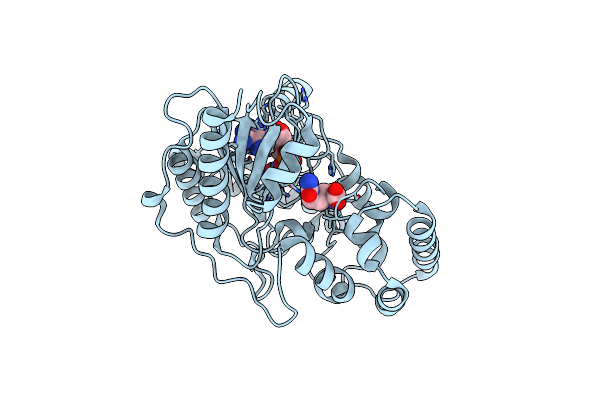 |
Crystal Structure Of Arginine Kinase In Complex With L-Citrulline And Mgadp
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2012-10-03 Classification: TRANSFERASE Ligands: MG, ADP, CIR |
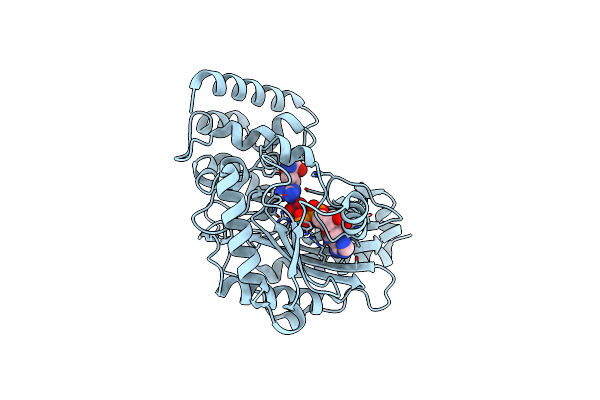 |
Crystal Structure Of Arginine Kinase In Complex With D-Arginine, Mgadp, And Nitrate.
Organism: Limulus polyphemus
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2012-10-03 Classification: TRANSFERASE Ligands: ADP, MG, NO3, DAR |

