Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Hippeastrum hybrid cultivar
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2022-10-05 Classification: SUGAR BINDING PROTEIN Ligands: BMA, PO4 |
 |
Crystal Structure Of Pi3K Alpha In Complex With 3-(2-Amino-Benzooxazol-5-Yl)-1-Isopropyl-1H-Pyrazolo[3,4-D]Pyrimidin-4-Ylamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: FE5 |
 |
Crystal Structure Of Pi3K Alpha In Complex With 3-(2-Amino-Benzooxazol-5-Yl)-1-Isopropyl-4-Methyl-1H-Pyrazolo[3,4-D]Pyrimidin-6-Ylamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: FCZ |
 |
Crystal Structure Of Pi3K Alpha In Complex With 3-(2-Amino-Benzooxazol-5-Yl)-4-Chloro-1-Isopropyl-1H-Pyrazolo[3,4-D]Pyrimidin-6-Ylamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.74 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: FDH |
 |
Crystal Structure Of Pi3K Alpha In Complex With 3-(2-Amino-Benzooxazol-5-Yl)-1-Isopropyl-1H-Pyrazolo[3,4-D]Pyrimidine-4,6-Diamine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2019-10-02 Classification: TRANSFERASE Ligands: FDW |
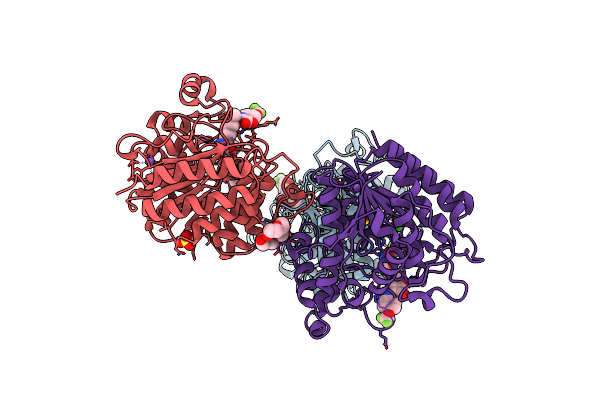 |
Crystal Structure Of Human Hdac2 In Complex With (R)-6-[3,4-Dioxo-2-(4-Trifluoromethoxy-Phenylamino)-Cyclobut-1-Enylamino]-Heptanoic Acid Hydroxyamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2018-06-27 Classification: HYDROLASE Ligands: ZN, CA, NA, EL8, 1PE, SO4, PG4 |
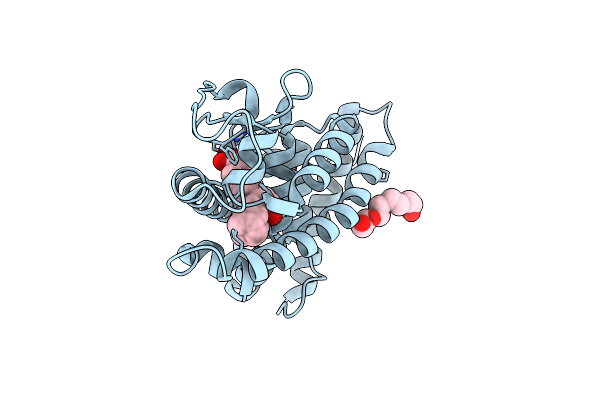 |
Structure-Based Design Of Trifarotene (Cd5789), A Potent And Selective Rar Gamma Agonist For The Treatment Of Acne
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-23 Classification: SIGNALING PROTEIN Ligands: E9T, PG4 |
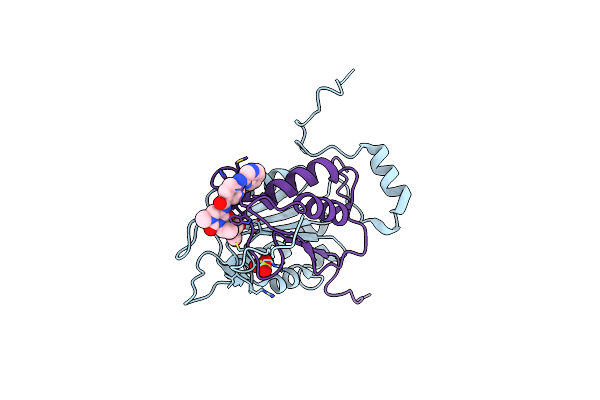 |
Crystal Structure Of Human Caspase-1 With N-{3-[1-((S)-2-Hydroxy-5-Oxo-Tetrahydro-Furan-3-Ylcarbamoyl)-Ethyl]-1-Methyl-2,4-Dioxo-1,2,3,4-Tetrahydro-Pyrimidin-5-Yl}-4-(Quinoxalin-2-Ylamino)-Benzamide
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2018-05-02 Classification: HYDROLASE Ligands: CVE, SO4 |
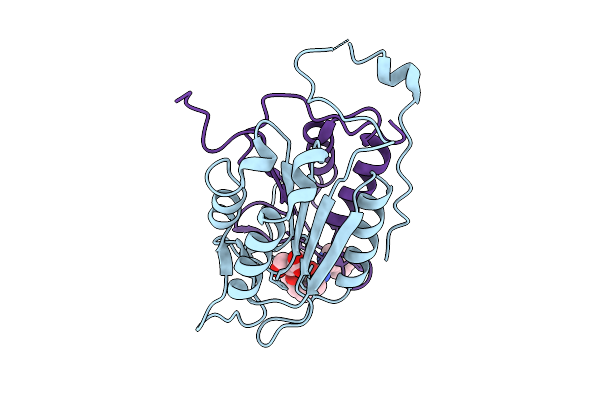 |
Crystal Structure Of Human Caspase-1 With (3S,6S,10As)-N-((2S,3S)-2-Hydroxy-5-Oxotetrahydrofuran-3-Yl)-6-(Isoquinoline-1-Carboxamido)-5-Oxodecahydropyrrolo[1,2-A]Azocine-3-Carboxamide (Pge-3935199)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Release Date: 2018-02-28 Classification: HYDROLASE Ligands: N7N |
 |
Crystal Structure Of Human Caspase-1 With 2-((2-Naphthoyl)-L-Valyl)-4-Hydroxy-N-((3S)-2-Hydroxy-5-Oxotetrahydrofuran-3-Yl)-2-Azabicyclo[2.2.2]Octane-3-Carboxamide (Compound 1)
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2017-12-20 Classification: HYDROLASE Ligands: WE0 |
 |
Discovery Of Phenoxyindazoles And Phenylthioindazoles As Rorg Inverse Agonists
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-11-16 Classification: TRANSCRIPTION Ligands: 79U |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-08-03 Classification: DNA BINDING PROTEIN Ligands: PO4, CL |
 |
Metallo-Beta-Lactamase Bcii Cd Substituted From Bacillus Cereus At 1.35 Angstroms Resolution
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2004-05-18 Classification: HYDROLASE Ligands: CD, CIT |
 |
Human Protein L-Isoaspartate O-Methyltransferase With S-Adenosyl Homocysteine
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2002-03-13 Classification: TRANSFERASE Ligands: SAH |
 |
Gadolinium Derivative Of Tetragonal Hen Egg-White Lysozyme At 1.7 A Resolution
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:1.72 Å Release Date: 2002-01-15 Classification: HYDROLASE Ligands: DO3, CL, GD |
 |
1,3-Alpha-1,4-Beta-D-Galactose-4-Sulfate- 3,6-Anhydro-D-Galactose-2-Sulfate 4 Galactohydrolase
Organism: Alteromonas sp. atcc43554
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2001-11-27 Classification: HYDROLASE Ligands: CA, CL, NA, GOL |
 |
Structure Of Mure The Udp-N-Acetylmuramyl Tripeptide Synthetase From E. Coli
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2001-09-13 Classification: LIGASE Ligands: CL, UAG, API |
 |
Organism: Gallus gallus
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2001-01-24 Classification: DNA BINDING PROTEIN Ligands: PO4, CL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2001-01-17 Classification: LIGASE Ligands: UMA |
 |
1,3-Alpha-1,4-Beta-D-Galactose-4-Sulfate-3,6-Anhydro-D-Galactose 4 Galactohydrolase
Organism: Pseudoalteromonas carrageenovora
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2001-01-16 Classification: HYDROLASE Ligands: CD, CL |