Search Count: 146
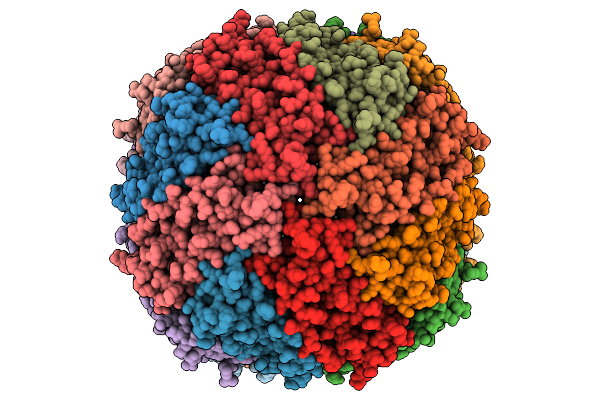 |
Cryo-Et Subtomogram-Averaged Structure Of Mouse Heavy-Chain Apoferritin Resolved At 2.71 Angstroms
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2026-01-28 Classification: OXIDOREDUCTASE Ligands: MG |
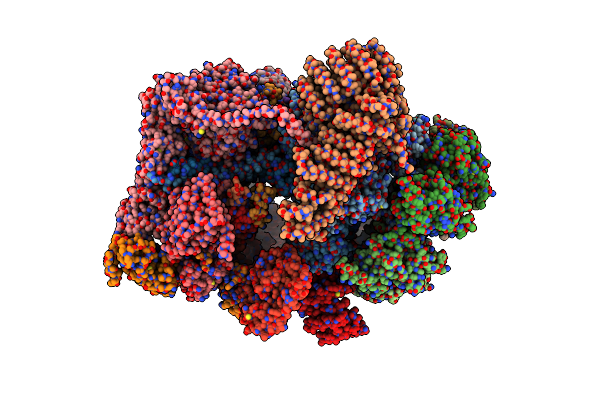 |
Cryo-Em Structure Of Human 26S Rp (Ed State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens, Aequorea victoria
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: VIRAL PROTEIN Ligands: NAG |
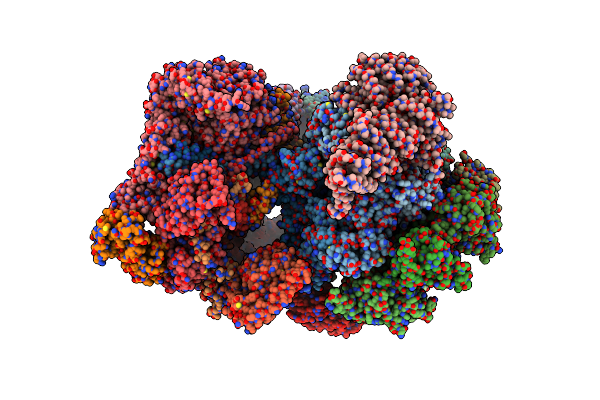 |
Cryo-Em Structure Of Human 26S Rp (Eb State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Four Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-25 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
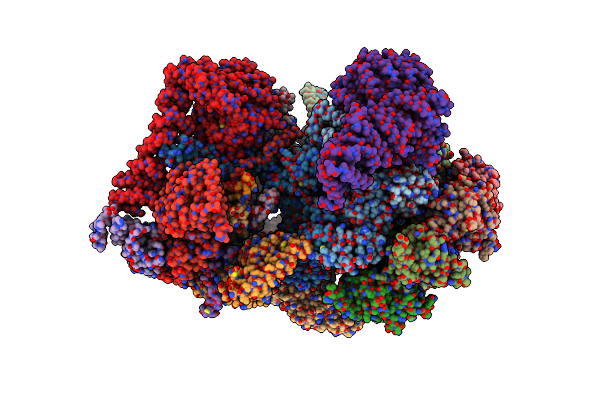 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Without Any Bound Substrate.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Cryo-Em Structure Of Human 26S Proteasomal Rp Subcomplex (Ea State) Bound To K11/K48-Branched Ubiquitin (Ub) Chain Composed Of Three Ub.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-12-18 Classification: CYTOSOLIC PROTEIN Ligands: ATP, MG, ADP |
 |
Organism: Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN Ligands: K |
 |
Organism: Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN |
 |
Organism: Sulfolobus acidocaldarius dsm 639
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2024-12-11 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Mouse Heavy-Chain Apoferritin Resolved At 1.51 Angstroms
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-21 Classification: OXIDOREDUCTASE Ligands: MG, K, FE |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-06-19 Classification: VIRAL PROTEIN Ligands: NAG |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2024-04-03 Classification: METAL TRANSPORT Ligands: ATP, TL |
 |
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2024-04-03 Classification: METAL BINDING PROTEIN Ligands: ATP, NA |
 |
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ADP, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Mgcl2
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Edta And Egta
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Edta And Egta, Vertical C2 Symmetry Axis
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Potassium Transporter Ktrab From Bacillus Subtilis In Atp-Bound State With Addition Of Edta And Egta, C1 Symmetry
Organism: Bacillus subtilis
Method: ELECTRON MICROSCOPY Release Date: 2024-04-03 Classification: TRANSPORT PROTEIN Ligands: ATP, NA, K |
 |
Dark, Fully Reduced Structure Of The Mmcpdii-Dna Complex As Produced At Swissfel
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: FAD, SO4 |
 |
Tr-Sfx Mmcpdii-Dna Complex: 100 Ps Snapshot. Includes 100Ps, Dark, And Extrapolated Structure Factors
Organism: Methanosarcina mazei, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2023-11-22 Classification: DNA BINDING PROTEIN Ligands: SO4, FAD |

