Search Count: 40
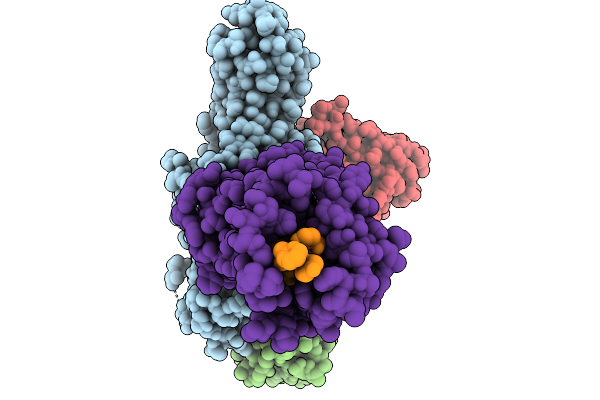 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 1)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
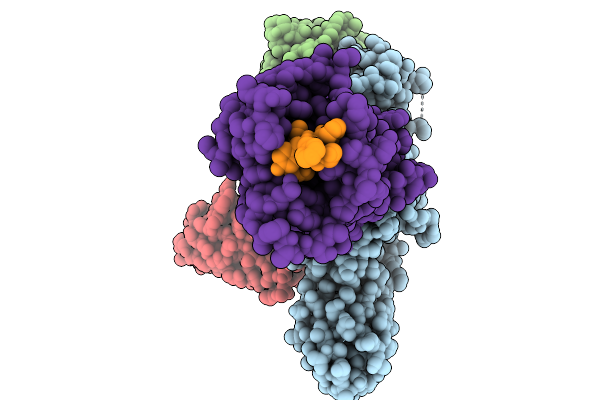 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 2)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
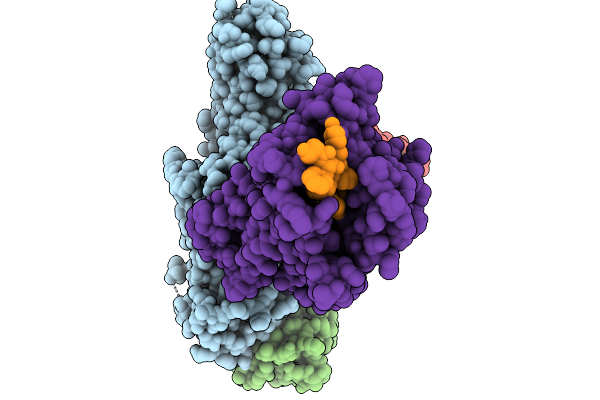 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 3)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
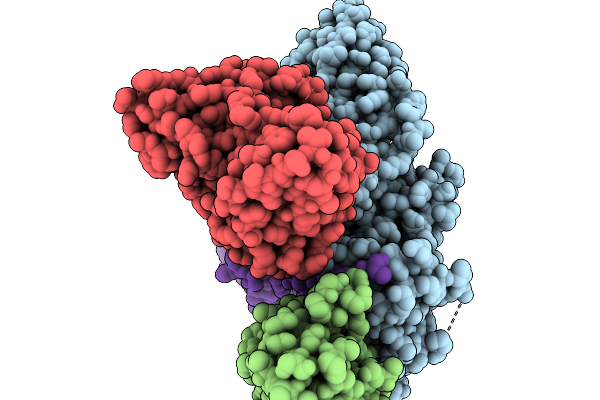 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 1 (Conformation 4)
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Composite Map Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Chemerin And Beta-Arrestin 2
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: Y01 |
 |
Cryo-Em Structure Of The G Protein-Coupled Receptor 1 (Gpr1) Bound To Beta-Arrestin 1 In Ligand-Free State
Organism: Homo sapiens, Escherichia phage ecszw-2
Method: ELECTRON MICROSCOPY Release Date: 2025-11-19 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM Ligands: PAM |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: VIRAL PROTEIN/RNA Ligands: ZN, CA, K5X |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: VIRAL PROTEIN/RNA Ligands: ZN, CA, EIF |
 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Resolution:2.90 Å Release Date: 2025-03-19 Classification: VIRAL PROTEIN/RNA Ligands: ZN, MG |
 |
Cryo-Em Structure Of The Chemokine-Like Receptor 1 In Complex With Chemerin And Gi1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
Cryo-Em Structure Of The G-Protein Coupled Receptor 1 (Gpr1) In Complex With Chemerin And Gi1
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN |
 |
Organism: Mus musculus, Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2025-03-12 Classification: MEMBRANE PROTEIN/IMMUNE SYSTEM |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-29 Classification: TRANSLOCASE/DNA Ligands: ANP |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSLOCASE |
 |
Organism: Thermus antranikianii dsm 12462
Method: ELECTRON MICROSCOPY Release Date: 2025-01-22 Classification: TRANSLOCASE/DNA Ligands: ANP |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: PLANT PROTEIN Ligands: A1D8A |
 |
Organism: Penaeus vannamei
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2023-12-13 Classification: LYASE Ligands: PEP, MG, SO4 |
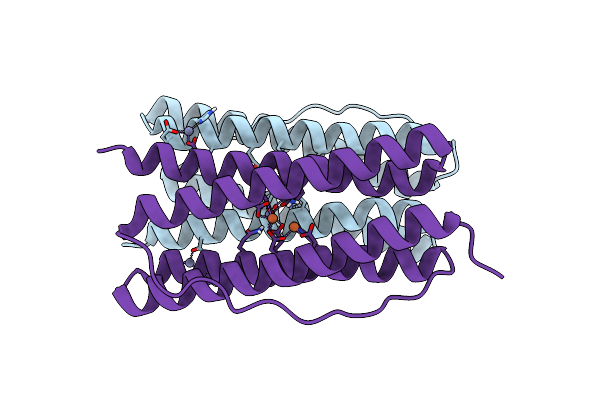 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-02-15 Classification: METAL BINDING PROTEIN Ligands: ZN, FE |
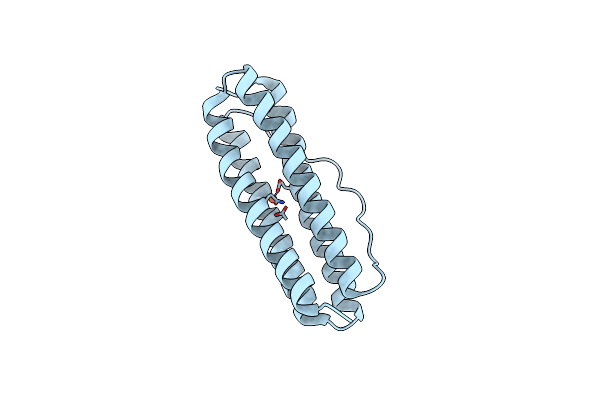 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-02-15 Classification: METAL BINDING PROTEIN Ligands: FE |
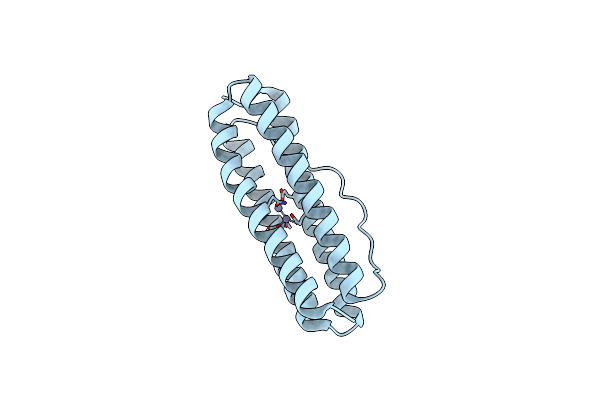 |
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2023-02-15 Classification: METAL BINDING PROTEIN Ligands: ZN |

