Search Count: 27
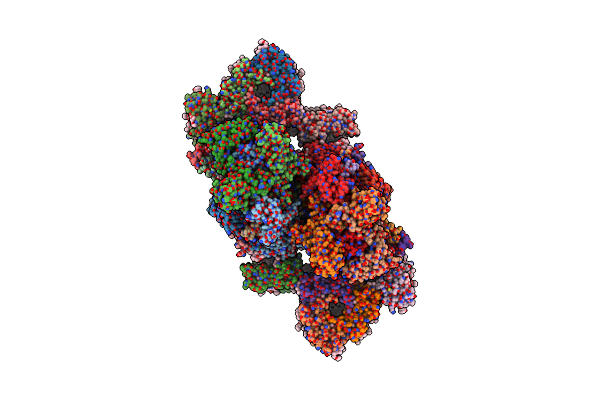 |
Cryo-Em Structure Of Spinach Psii-Lhcii Supercomplex At 3.2 Angstrom Resolution
|
 |
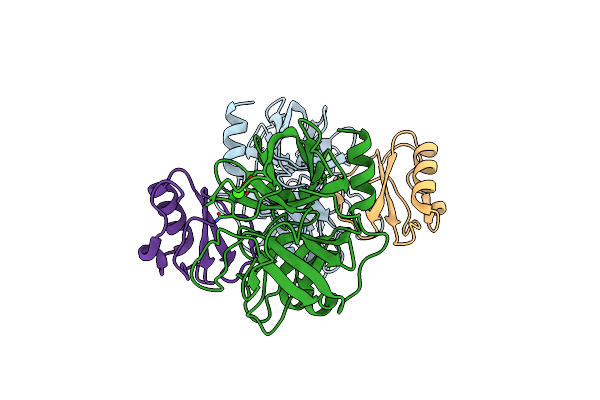
Crystal Structure Of Buckwheat Trypsin Inhibitor Rbti At 1.84 Angstrom Resolution
Organism: Fagopyrum esculentum
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2011-07-06 Classification: HYDROLASE INHIBITOR |
 |
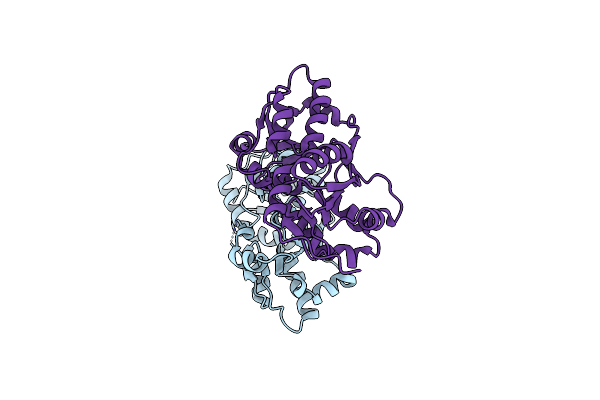
Organism: Fagopyrum esculentum, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2011-07-06 Classification: HYDROLASE/HYDROLASE INHIBITOR Ligands: CA |
 |
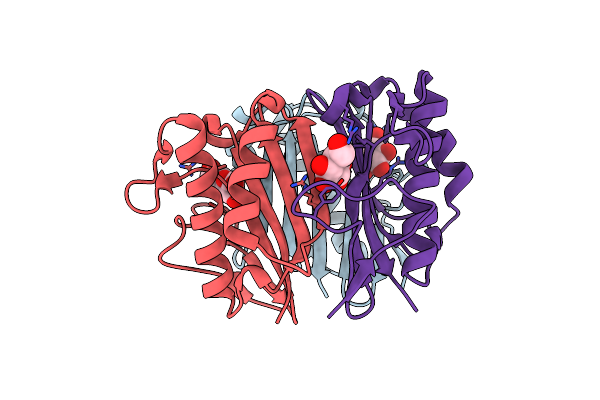
Crystal Structure Of Uroporphyrinogen Iii Synthase From Pseudomonas Syringae Pv. Tomato Dc3000
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-06-22 Classification: LYASE |
 |
Crystal Structure Of Pspto -Psp Protein In Complex With D-Beta-Glucose From Pseudomonas Syringae Pv. Tomato Str. Dc3000
Organism: Pseudomonas syringae pv. tomato
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2010-06-16 Classification: SUGAR BINDING PROTEIN Ligands: BGC |
 |
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2007-04-10 Classification: ISOMERASE Ligands: SO4, ZN, GOL |
 |
Crystal Structure Of The Mycothiol-Dependent Maleylpyruvate Isomerase H52A Mutant
Organism: Corynebacterium glutamicum
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2007-04-10 Classification: ISOMERASE Ligands: SO4, GOL |
 |
Crystal Structure Of C-Terminal Desundecapeptide Nitrite Reductase From Achromobacter Cycloclastes
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-12-20 Classification: OXIDOREDUCTASE Ligands: CU, CL |
 |
Crystal Structure Of Human Sulfotransferase Sult1A3 In Complex With Dopamine And 3-Phosphoadenosine 5-Phosphate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2005-08-30 Classification: TRANSFERASE Ligands: A3P, LDP |
 |
Crystal Structure Of Earthworm Fibrinolytic Enzyme Component B: A Novel, Glycosylated Two-Chained Trypsin
Organism: Eisenia fetida
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2005-04-19 Classification: HYDROLASE Ligands: MG, SO4 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-03-22 Classification: UNKNOWN FUNCTION |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.42 Å Release Date: 2004-12-28 Classification: HORMONE/GROWTH FACTOR |
 |
Organism: Solanum tuberosum
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-06-29 Classification: METAL BINDING PROTEIN Ligands: CA, MPD |
 |
Crystal Structure Of C-Terminal Despentapeptide Nitrite Reductase From Achromobacter Cycloclastes At Ph6.2
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: CU, SO4, MES |
 |
Crystal Structure Of C-Terminal Despentapeptide Nitrite Reductase From Achromobacter Cycloclastes At Ph5.0
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-03-30 Classification: OXIDOREDUCTASE Ligands: CU, SO4, ACY |
 |
Crystal Structure Of A No-Forming Nitrite Reductase Mutant: An Analog Of A Transition State In Enzymatic Reaction
Organism: Achromobacter cycloclastes
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2003-11-04 Classification: OXIDOREDUCTASE Ligands: CU |
 |
Organism: Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2003-02-11 Classification: TRANSFERASE |
 |
Organism: Ovis aries
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2003-02-11 Classification: TRANSFERASE Ligands: ZN, K, ATP |
 |
Organism: Desulfovibrio vulgaris
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2002-09-11 Classification: ELECTRON TRANSPORT Ligands: FE, ZN |
 |
Crystal Structure Of Manganese Superoxide Dismutases From Bacillus Halodenitrificans
Organism: Virgibacillus halodenitrificans
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2002-08-14 Classification: OXIDOREDUCTASE Ligands: MN, ZN |

