Search Count: 283
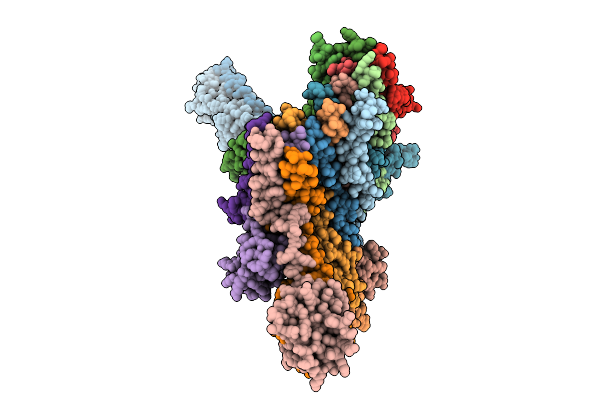 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Lacking The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
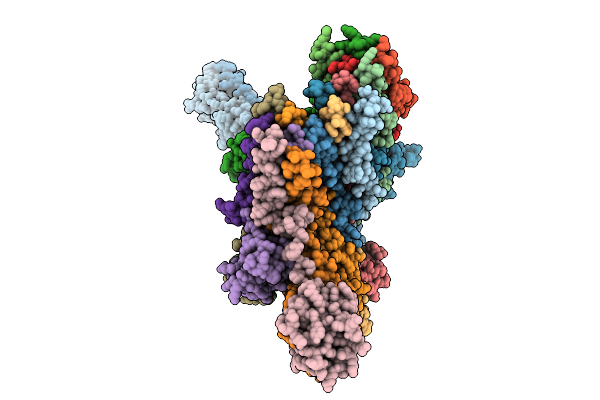 |
Cryo-Em Structure Of The Vaccinia Virus Entry/Fusion Complex (Efc) Including The F9 Subunit
Organism: Orthopoxvirus vaccinia
Method: ELECTRON MICROSCOPY Release Date: 2025-12-17 Classification: VIRAL PROTEIN |
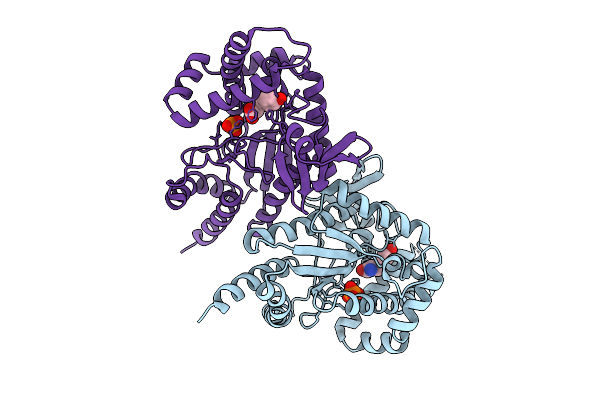 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, TYR |
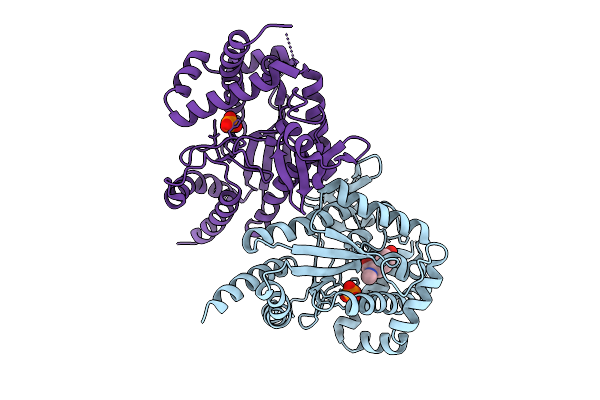 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PO4, I3J |
 |
Organism: Photorhabdus luminescens
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: TRANSFERASE Ligands: PG4, A1A46, PO4, EDO, PG0, PG6 |
 |
Organism: Streptomyces lavendulae
Method: X-RAY DIFFRACTION Release Date: 2025-06-18 Classification: OXIDOREDUCTASE Ligands: FE, ZN, GOL |
 |
Organism: Pseudooceanicola atlanticus
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2025-04-09 Classification: UNKNOWN FUNCTION |
 |
Organism: Pseudooceanicola atlanticus
Method: X-RAY DIFFRACTION Resolution:2.27 Å Release Date: 2025-04-09 Classification: UNKNOWN FUNCTION |
 |
Organism: Streptomyces sp. cfmr 7
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: ACY, MG, EDO |
 |
Crystal Structure Of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed With Manganese (Ii), 2-Oxoglutarate And 6-Nitronorleucine
Organism: Streptomyces sp. cfmr 7
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: MN, AKG, 6HN, IOD, EDO, GOL |
 |
Crystal Structure Of Hrmj From Streptomyces Sp. Cfmr 7 (Hrmj-Ssc) Complexed With Vanadyl(Iv)-Oxo, Succinate And 6-Nitronorleucine
Organism: Streptomyces sp. cfmr 7
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: 6HN, VVO, SIN, GOL, BR |
 |
Organism: Streptomyces sp. ag109_g2-6
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: K, SCN, GOL |
 |
Crystal Structure Of Hrmj From Streptomyces Sp. Ag109_G2-6 (Hrmj-Ssa) Complexed With Ferric Iron(Iii) And 2-Oxoglutarate
Organism: Streptomyces sp. ag109_g2-6
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: AKG, FE |
 |
Crystal Structure Of Hrmj From Streptomyces Sp. Ag109_G2-6 (Hrmj-Ssa) Complexed With Vanadyl(Iv)-Oxo And Succinate
Organism: Streptomyces sp. ag109_g2-6
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2025-02-12 Classification: OXIDOREDUCTASE Ligands: SIN, VVO |
 |
Organism: Dragon grouper nervous necrosis virus
Method: SOLUTION NMR Release Date: 2024-10-30 Classification: VIRAL PROTEIN |
 |
Structure Of Calcium-Sensing Receptor In Complex With Positive Allosteric Modulator '6218
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: NAG, CA, TRP, PO4, A1ATP |
 |
Structure Of Calcium-Sensing Receptor In Complex With Positive Allosteric Modulator '54149
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2024-10-02 Classification: MEMBRANE PROTEIN Ligands: NAG, CA, TRP, PO4, A1ATX |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph8.0 (3.23A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph6.5 (2.82A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |
 |
Cryo-Em Structure Of Dragon Grouper Nervous Necrosis Virus-Like Particle At Ph5.0 (3.52A)
Organism: Dragon grouper nervous necrosis virus
Method: ELECTRON MICROSCOPY Release Date: 2024-08-14 Classification: VIRUS Ligands: CA |

