Search Count: 33
 |
Organism: Acinetobacter genomosp. 16bj
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: CELL ADHESION |
 |
Organism: Bacteria abnormis, Acinetobacter phage ap205
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS/RNA |
 |
Organism: Acinetobacter phage ap205, Acinetobacter genomosp. 16bj
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
 |
Organism: Acinetobacter phage ap205, Acinetobacter genomosp. 16bj
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS |
 |
Organism: Acinetobacter phage ap205
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Acinetobacter phage ap205
Method: ELECTRON MICROSCOPY Release Date: 2024-03-06 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-10-11 Classification: RNA BINDING PROTEIN Ligands: SO4, NH2, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-09-06 Classification: GENE REGULATION |
 |
Organism: Homo sapiens, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-07-26 Classification: SIGNALING PROTEIN Ligands: GDP, AF3, MG |
 |
Organism: Escherichia phage qbeta
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia phage qbeta
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia phage qbeta
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia phage qbeta
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRUS LIKE PARTICLE |
 |
Organism: Escherichia virus qbeta, Escherichia phage qbeta
Method: ELECTRON MICROSCOPY Release Date: 2022-01-26 Classification: VIRUS/RNA |
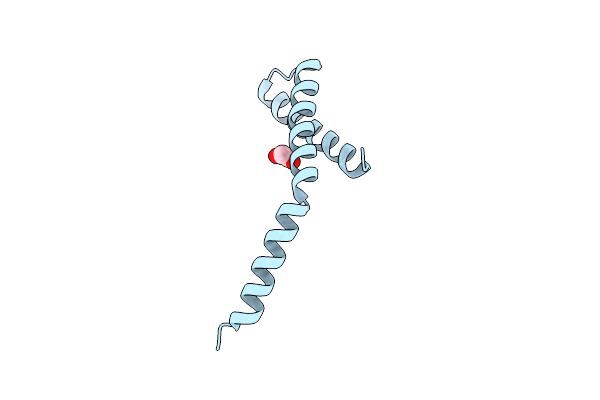 |
Organism: Enterobacteria phage t4
Method: X-RAY DIFFRACTION Resolution:2.21 Å Release Date: 2020-06-24 Classification: VIRAL PROTEIN Ligands: EDO |
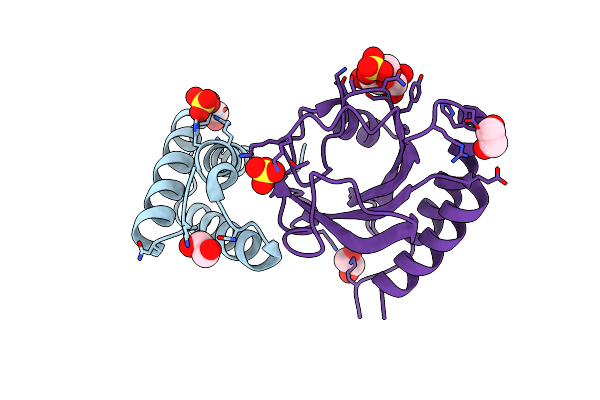 |
Crystal Structure Of The Complex Between Periplasmic Domains Of Antiholin Ri And Holin T From T4 Phage, In P6522
Organism: Escherichia phage ecml-134, Escherichia phage vb_ecom_nbg2
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-06-24 Classification: VIRAL PROTEIN Ligands: SO4, EDO, BTB, CL |
 |
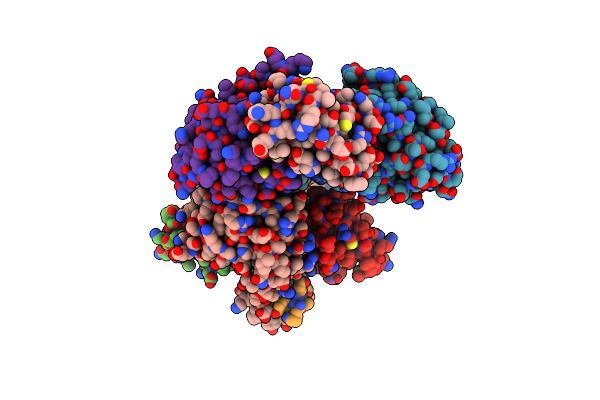
Crystal Structure Of The Complex Between Periplasmic Domains Of Antiholin Ri And Holin T From T4 Phage, In H32
Organism: Escherichia phage ecml-134, Escherichia phage vb_ecom_nbg2
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2020-06-24 Classification: VIRAL PROTEIN |
 |
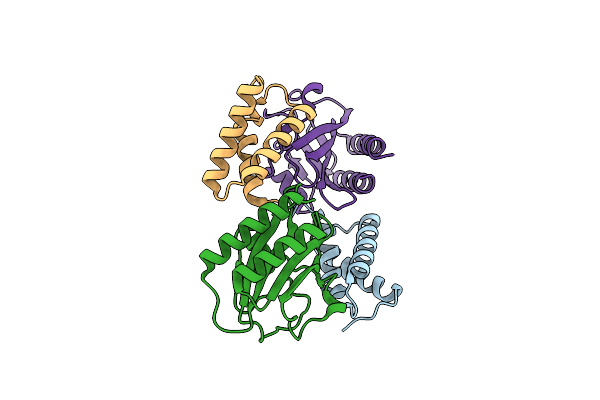
Crystal Structure Of The Complex Between Periplasmic Domains Of Antiholin Ri And Holin T From T4 Phage, In P21
Organism: Enterobacteria phage t4, Escherichia phage vb_ecom_nbg2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-06-24 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of Indoleamine 2,3-Dioxygenagse 1 (Ido1) In Complex With Compound 47
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2020-03-25 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: HEM, DO9 |
 |
Crystal Structure Of Indoleamine 2,3-Dioxygenagse 1 (Ido1) In Complex With Compound 36
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2020-03-25 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: HEM, DU6 |

