Search Count: 9
 |
Predicting Amino Acid Preferences In The Complementarity Determining Regions Of An Antibody-Antigen Recognition Interface
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2012-02-22 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2012-02-15 Classification: TRANSFERASE/TRANSFERASE INHIBITOR Ligands: 0KD |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2011-11-16 Classification: HYDROLASE |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-04-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, TCL |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2011-04-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, TCL |
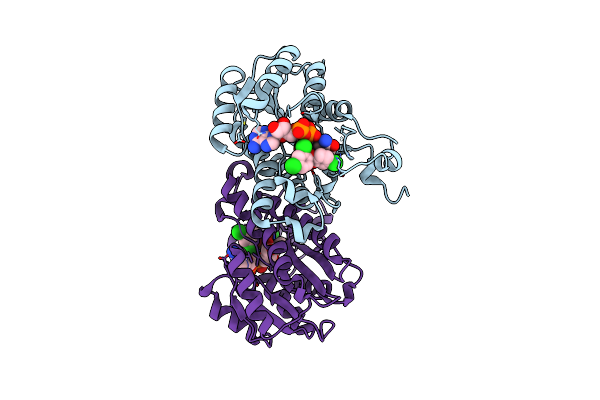 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2011-04-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: NAD, TCL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2004-02-17 Classification: TRANSFERASE Ligands: SO4, AOX |
 |
Solution Structure And Backbone Dynamics Of A Double Mutant Single-Chain Monellin(Scm) Determined By Nuclear Magnetic Resonance Spectroscopy
Organism: Dioscoreophyllum cumminsii
Method: SOLUTION NMR Release Date: 2001-06-06 Classification: PLANT PROTEIN |
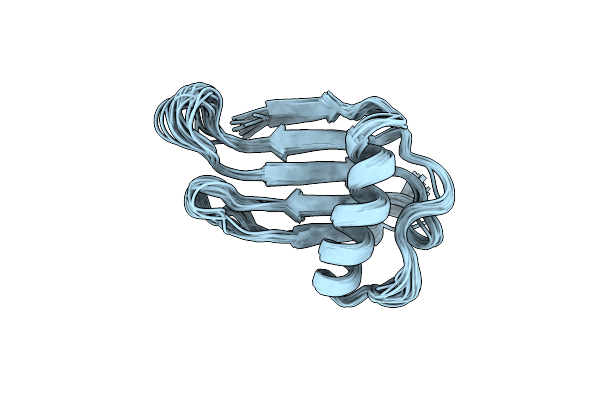 |
High-Resolution Solution Structure Of A Sweet Protein Single-Chain Monellin (Scm) Determined By Nuclear Magnetic Resonance Spectroscopy And Dynamical Simulated Annealing Calculations, 21 Structures
Organism: Dioscoreophyllum cumminsii
Method: SOLUTION NMR Release Date: 1999-06-08 Classification: SWEET PROTEIN |

