Search Count: 22
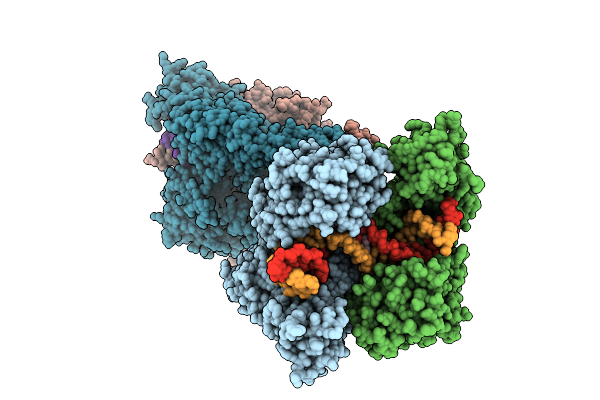 |
Crystal Structure Of Topoisomerase Iv From Klebsiella Pneumoniae In Complex With Dna And Bwc0977, A Dual-Targeting Broad-Spectrum Novel Bacterial Topoisomerase Inhibitor.
Organism: Klebsiella pneumoniae subsp. pneumoniae mgh 78578, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-11-12 Classification: ISOMERASE/DNA/ISOMERASE INHIBITOR Ligands: A1L5V |
 |
Crystal Structure Of Apobec3G Catalytic Domain Complex With Ssdna Containing 2'-Deoxy Zebularine.
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2022-12-14 Classification: HYDROLASE/DNA Ligands: ZN |
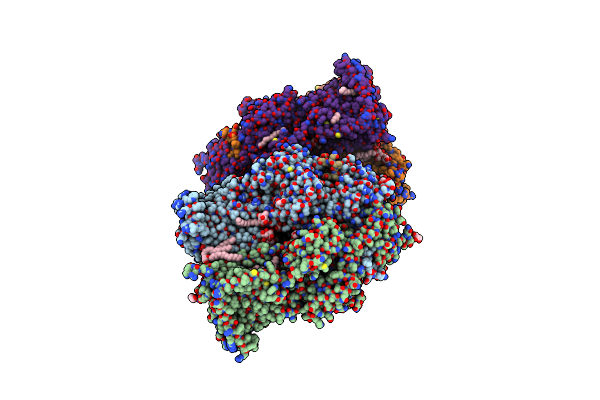 |
Crystal Structure Of Multidrug Efflux Transporter Oqxb From Klebsiella Pneumoniae
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2021-09-22 Classification: MEMBRANE PROTEIN Ligands: LMT, PTY, GOL |
 |
Crystal Structure Of A Soluble Variant Of Full-Length Human Apobec3G (Ph 7.6)
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-12-23 Classification: HYDROLASE/DNA Ligands: ZN, CL, GOL |
 |
Crystal Structure Of A Soluble Variant Of Full-Length Human Apobec3G (Ph 8.0)
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.02 Å Release Date: 2020-12-23 Classification: HYDROLASE/DNA Ligands: ZN |
 |
Crystal Structure Of A Soluble Variant Of Full-Length Human Apobec3G (Ph 9.0)
Organism: Homo sapiens, Escherichia coli
Method: X-RAY DIFFRACTION Resolution:3.49 Å Release Date: 2020-12-23 Classification: HYDROLASE/DNA Ligands: ZN |
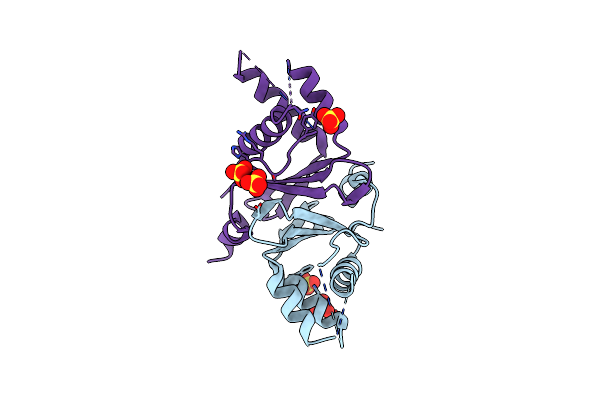 |
Organism: Schizosaccharomyces pombe 972h-
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-02-13 Classification: RNA BINDING PROTEIN Ligands: SO4 |
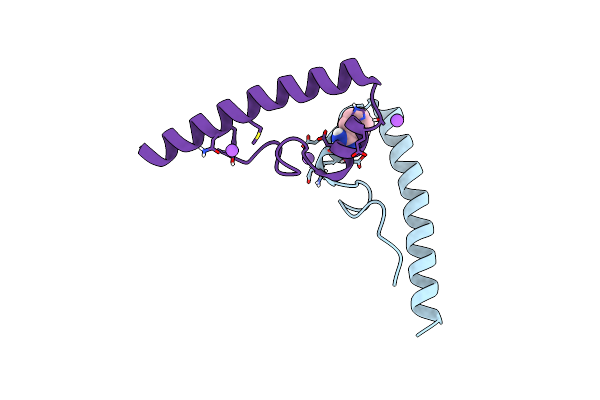 |
Crystal Structure Of The S. Cerevisiae Rtf1 Histone Modification Domain Mutant R126A
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c)
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2016-11-09 Classification: TRANSCRIPTION |
 |
Crystal Structure Of The S. Cerevisiae Rtf1 Histone Modification Domain Mutant R124A R126A R128A
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2016-10-26 Classification: TRANSCRIPTION |
 |
Organism: Saccharomyces cerevisiae, Xenopus laevis
Method: X-RAY DIFFRACTION Resolution:6.70 Å Release Date: 2016-08-03 Classification: DNA BINDING PROTEIN |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: NAD, MG, HRW |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: NAD, MG, KI5 |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: NAD, MG, 1PE, A0C |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: NAD, UUD, ACT |
 |
Organism: Mycobacterium tuberculosis
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2015-09-30 Classification: OXIDOREDUCTASE Ligands: NAD, KXU |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2015-08-26 Classification: GENE REGULATION Ligands: SO4 |
 |
Atomic Model For The N-Terminus Of Trao Fitted In The Full-Length Structure Of The Bacterial Pkm101 Type Iv Secretion System Core Complex
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:12.40 Å Release Date: 2013-04-03 Classification: MEMBRANE PROTEIN |
 |
Fitting Result In The O-Layer Of The Subnanometer Structure Of The Bacterial Pkm101 Type Iv Secretion System Core Complex Digested With Elastase
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:8.50 Å Release Date: 2013-04-03 Classification: CELL ADHESION |
 |
Fitting Results In The I-Layer Of The Subnanometer Structure Of The Bacterial Pkm101 Type Iv Secretion System Core Complex Digested With Elastase
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Resolution:8.50 Å Release Date: 2013-04-03 Classification: CELL ADHESION |
 |
Crystal Structure Of The Outer Membrane Complex Of A Type Iv Secretion System
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2009-12-01 Classification: TRANSPORT PROTEIN Ligands: MPD, LDA |

