Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
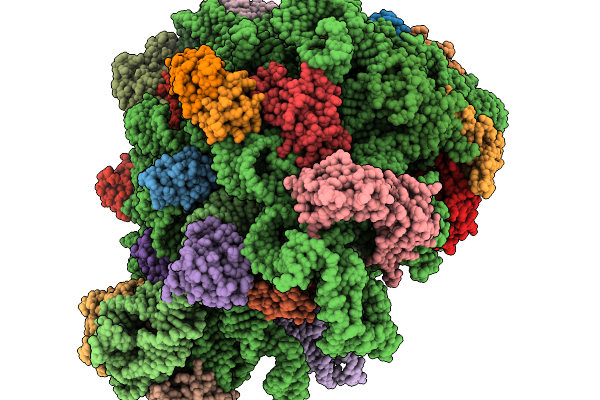 |
Structure Of Wt E.Coli Ribosome With Complexed Filament Nascent Chain At Length 47, With P-Site Trna
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
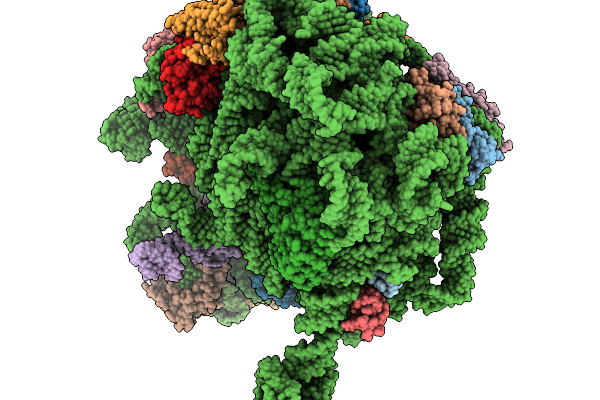 |
Structure Of Wt E.Coli Ribosome With Complexed Filament Nascent Chain At Length 31, With P-Site Trnas
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2026-01-14 Classification: RIBOSOME |
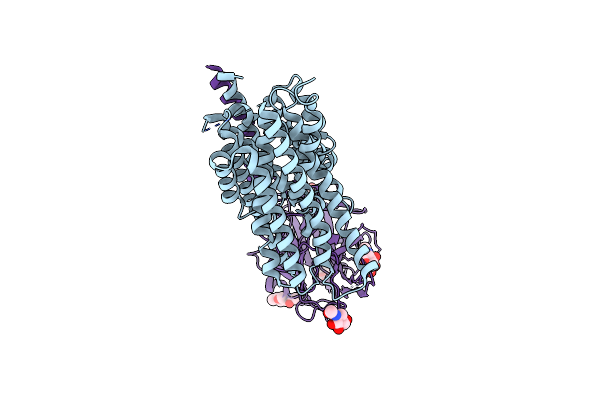 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2024-05-01 Classification: MEMBRANE PROTEIN Ligands: NAG |
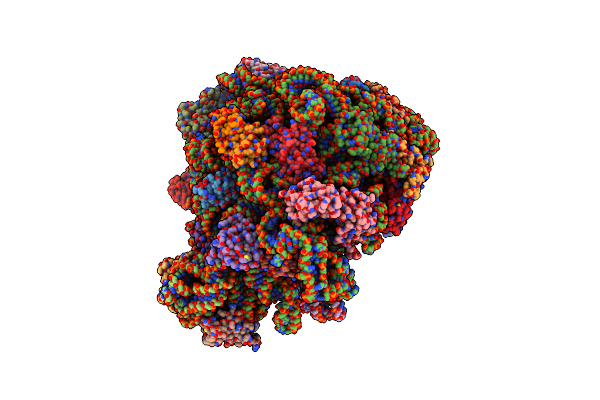 |
70S E. Coli Ribosome With An Extended Ul23 Loop From Candidatus Marinimicrobia And A Stalled Filamin Domain 5 Nascent Chain
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: RIBOSOME |
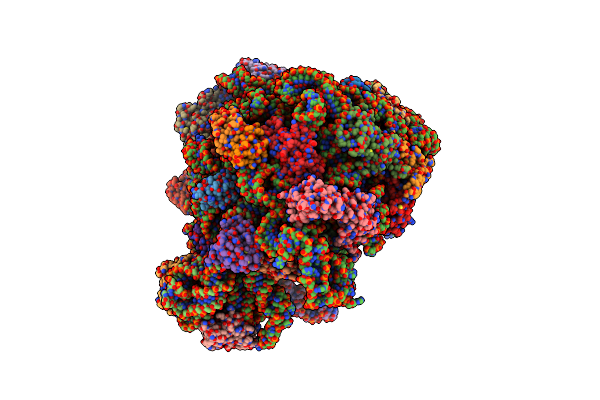 |
70S E. Coli Ribosome With An Extended Ul23 Loop From Candidatus Marinimicrobia
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: RIBOSOME |
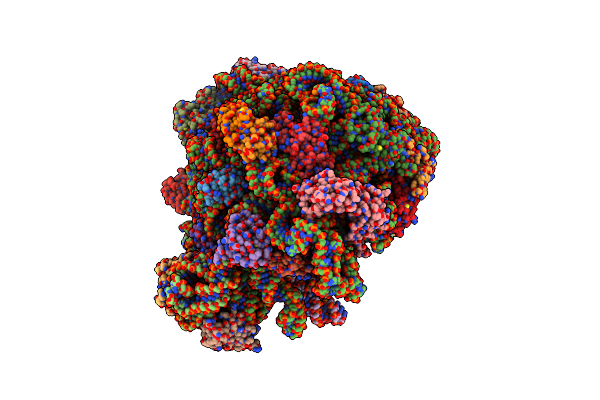 |
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: RIBOSOME |
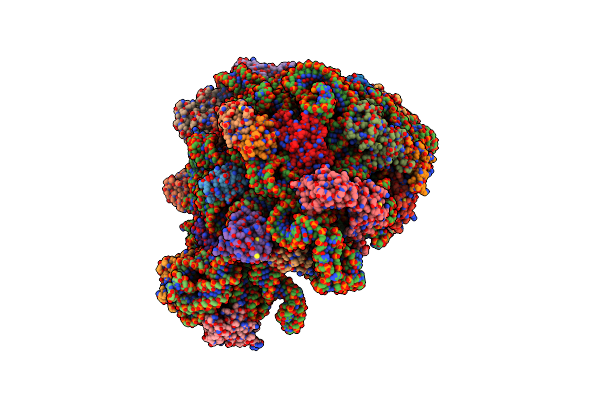 |
Organism: Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: RIBOSOME |
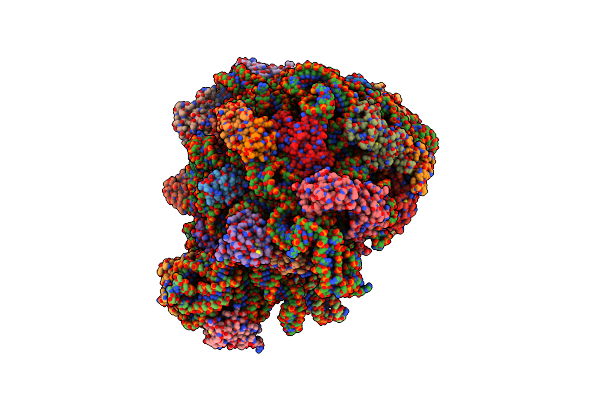 |
70S E. Coli Ribosome With Truncated Ul23 And Ul24 Loops And A Stalled Filamin Domain 5 Nascent Chain
Organism: Dictyostelium discoideum, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2022-08-10 Classification: RIBOSOME |
 |
Structure Of Sars-Cov-2 Nsp16/Nsp10 Complex In Presence Of Cap-1 Analog (M7Gpppamu) And Sah
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2021-05-05 Classification: HYDROLASE Ligands: MES, MG, SAH, YG4, EDO, ZN |
 |
Structure Of Sars-Cov-2 Nsp16/Nsp10 Complex In Presence Of S-Adenosyl-L-Homocysteine (Sah)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2021-05-05 Classification: HYDROLASE Ligands: MES, ACT, EDO, SAH, ZN |
 |
Structure Of Sars-Cov-2 Nsp16/Nsp10 In Complex With Rna Cap Analogue (M7Gpppa) And S-Adenosylmethionine
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2020-05-06 Classification: RNA BINDING PROTEIN Ligands: SAM, ADN, GTA, ZN, EDO |
 |
Organism: Dictyostelium discoideum
Method: SOLUTION NMR Release Date: 2019-04-10 Classification: STRUCTURAL PROTEIN |
 |
Structural Basis Of Asymmetric Dna Methylation And Atp-Triggered Long-Range Diffusion By Ecop15I
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2015-07-29 Classification: Hydrolase-DNA complex Ligands: MN, AMP, CA |
 |
Organism: Deinococcus radiophilus
Method: X-RAY DIFFRACTION Resolution:2.33 Å Release Date: 2014-05-14 Classification: HYDROLASE |
 |
Three-Dimensional Structure Of Dengue Virus Serotype 1 Complexed With 2 Hmab 14C10 Fab
Organism: Dengue virus 1, Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:7.00 Å Release Date: 2013-10-16 Classification: VIRUS |
 |
Three-Dimensional Structure Of Dengue Virus Serotype 1 Complexed With Hmab 14C10 Fab
|
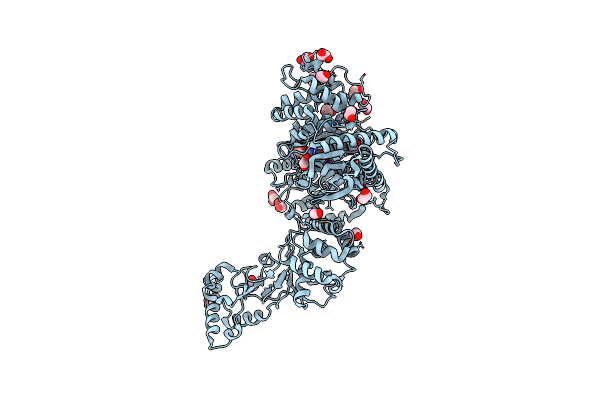 |
Characterization And Crystal Structure Of The Type Iig Restriction Endonuclease Bpusi
Organism: Bacillus pumilus
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2011-07-13 Classification: HYDROLASE, TRANSFERASE Ligands: SAH, EDO, MN, IOD |
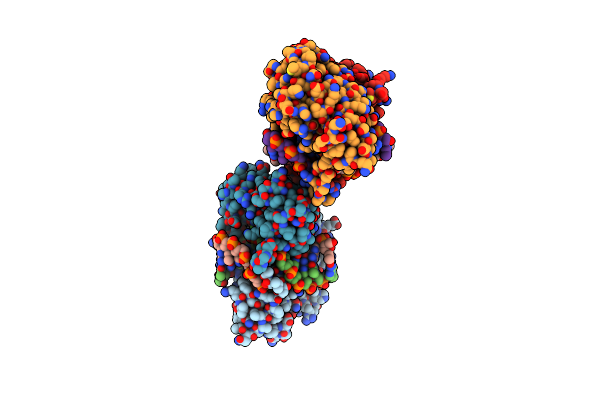 |
Organism: Oceanobacter kriegii
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2010-10-06 Classification: HYDROLASE/DNA Ligands: CA |
 |
An Extraordinary Mechanism Of Dna Perturbation Exhibited By The Rare-Cutting Hnh Restriction Endonuclease Paci
Organism: Pseudomonas alcaligenes
Method: X-RAY DIFFRACTION Resolution:1.97 Å Release Date: 2010-04-21 Classification: HYDROLASE/DNA Ligands: ZN, CA |
 |
Organism: Pseudomonas alcaligenes
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2010-04-21 Classification: HYDROLASE/DNA Ligands: ZN, MG, PT, NA, EDO, SO4 |