Search Count: 16
 |
Crystal Structure Of Periplasmic Domain Of Helicobacter Pylori Flil (Residues 81 To 183) (Crystal Form A)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-03-23 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of Periplasmic Domain Of Helicobacter Pylori Flil (Residues 81 To 183) (Crystal Form B)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2022-03-23 Classification: MOTOR PROTEIN |
 |
Crystal Structure Of Periplasmic Domain Of Helicobacter Pylori Flil (Residues 81 To 183) (Crystal Form C)
Organism: Helicobacter pylori
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-03-23 Classification: MOTOR PROTEIN |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2019-12-25 Classification: antitumor protein/inhibitor Ligands: ZN, GOL, JAV |
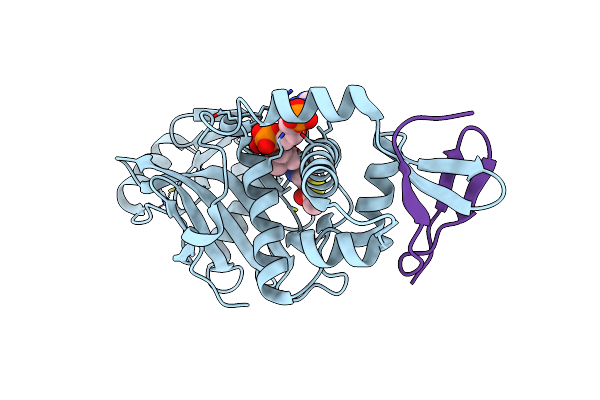 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.13 Å Release Date: 2019-12-25 Classification: antitumor protein/transferase Ligands: ZN, ACO |
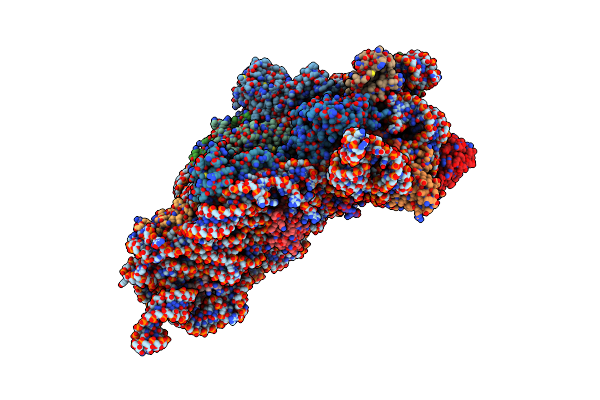 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With An Unmodifed Anticodon Stem Loop (Asl) Of Escherichia Coli Transfer Rna Arginine 2 (Trnaarg2) Bound To An Mrna With An Cgu-Codon In The A-Site And Paromomycin
Organism: Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.54 Å Release Date: 2019-04-24 Classification: RIBOSOME Ligands: PAR, MG |
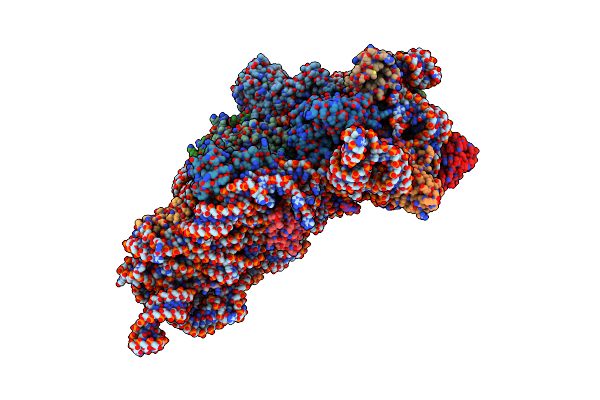 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With An Inosine (I34) Modified Anticodon Stem Loop (Asl) Of Escherichia Coli Transfer Rna Arginine 2 (Trnaarg2) Bound To An Mrna With An Cgu-Codon In The A-Site And Paromomycin
Organism: Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.46 Å Release Date: 2019-04-24 Classification: RIBOSOME Ligands: PAR, MG |
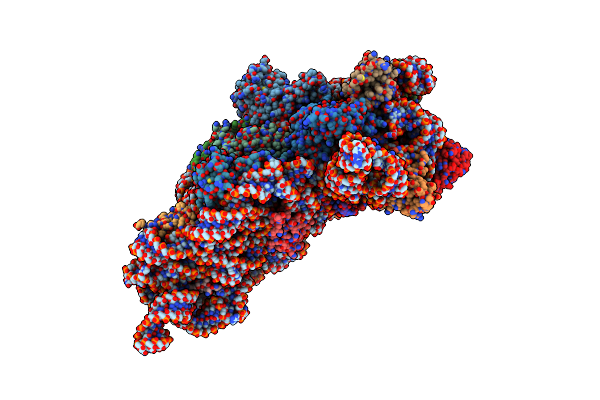 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A 2-Thiocytidine (S2C32) And Inosine (I34) Modified Anticodon Stem Loop (Asl) Of Escherichia Coli Transfer Rna Arginine 1 (Trnaarg1) Bound To An Mrna With An Cgc-Codon In The A-Site And Paromomycin
Organism: Escherichia coli, Thermus thermophilus hb8 (strain hb8 / atcc 27634 / dsm 579), Thermus thermophilus (strain hb8 / atcc 27634 / dsm 579)
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2019-04-24 Classification: RIBOSOME Ligands: PAR, MG, ZN |
 |
Structure Of The Thermus Thermophilus 30S Ribosomal Subunit Complexed With A 2-Thiocytidine (S2C32) And Inosine (I34) Modified Anticodon Stem Loop (Asl) Of Escherichia Coli Transfer Rna Arginine 1 (Trnaarg1) Bound To An Mrna With An Cgu-Codon In The A-Site And Paromomycin
Organism: Escherichia coli, Thermus thermophilus hb8
Method: X-RAY DIFFRACTION Resolution:3.33 Å Release Date: 2019-04-24 Classification: RIBOSOME Ligands: MG, PAR, ZN |
 |
Organism: Escherichia coli, Schizosaccharomyces pombe
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-07-01 Classification: SUGAR BINDING PROTEIN Ligands: SO4, MLI |
 |
Organism: Escherichia coli k-12
Method: SOLUTION NMR Release Date: 2015-06-17 Classification: TRANSFERASE |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2014-08-20 Classification: CELL CYCLE |
 |
Organism: Zygosaccharomyces rouxii
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-08-20 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2012-06-27 Classification: PROTEIN BINDING |
 |
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2008-02-19 Classification: CELL CYCLE, HYDROLASE Ligands: 3CN |
 |
Organism: Saccharomyces cerevisiae, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2008-02-19 Classification: CELL CYCLE, HYDROLASE Ligands: 3CN |

