Search Count: 20
 |
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With 5,6-Dihydrouridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:2.29 Å Release Date: 2014-01-22 Classification: LYASE Ligands: H2U, CL |
 |
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With 6-Azauridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2013-12-25 Classification: LYASE Ligands: UP6, CL, EDO, GOL, MG |
 |
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With Uridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2013-12-18 Classification: LYASE Ligands: U5P, GOL, EDO |
 |
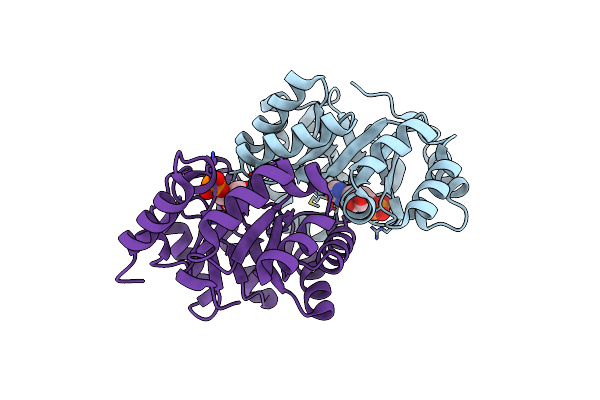
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-06-23 Classification: LYASE |
 |
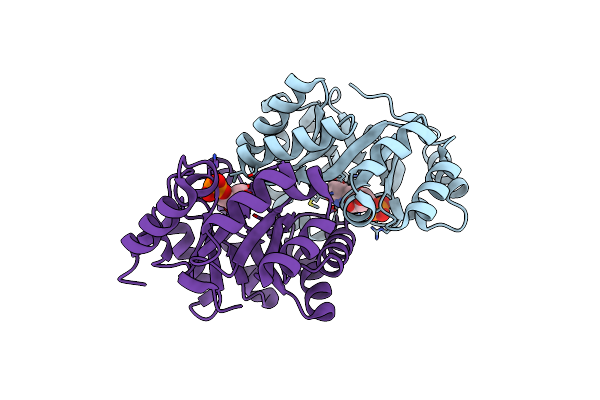
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With 6-Azauridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-06-23 Classification: LYASE Ligands: UP6 |
 |
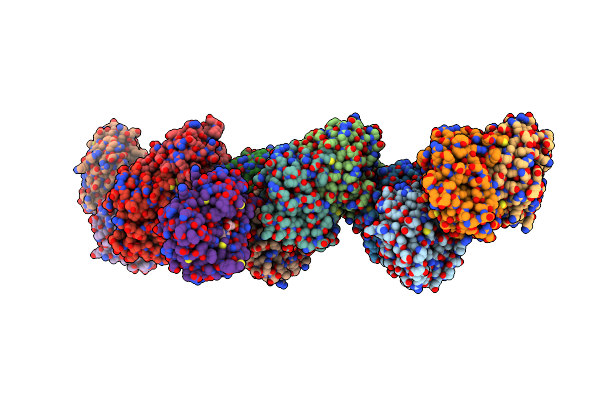
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With Uridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-06-23 Classification: LYASE Ligands: U5P |
 |
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With 5,6-Dihydroorotidine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2009-06-23 Classification: LYASE Ligands: 2OM |
 |
Crystal Structure Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With 5,6-Dihydrouridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2009-06-23 Classification: LYASE Ligands: H2U |
 |
Crystal Structure Of The Mutant D70G Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-06-23 Classification: LYASE |
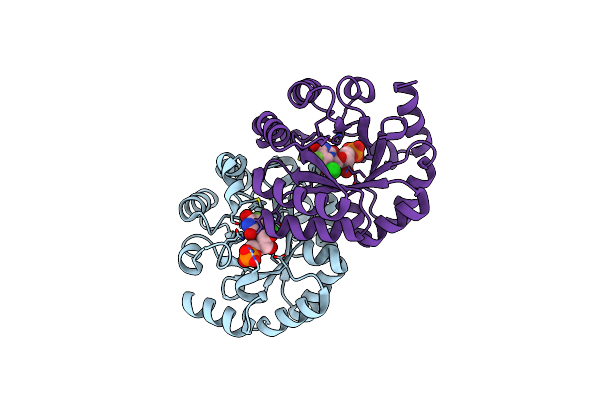 |
Crystal Structure Of The Mutant D70G Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With 5-Fluorouridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2009-06-23 Classification: LYASE Ligands: 5FU, CL |
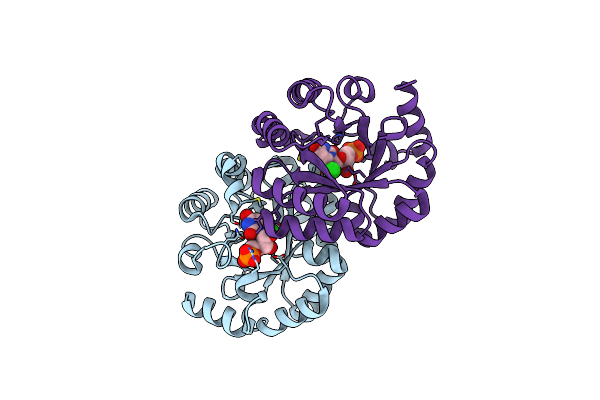 |
Crystal Structure Of The Mutant D70G Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With Uridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2009-06-23 Classification: LYASE Ligands: U5P, CL |
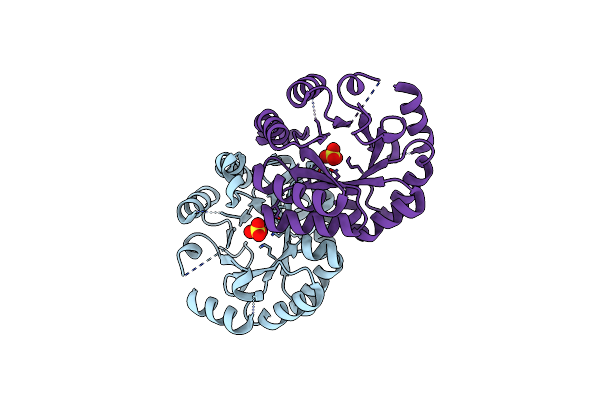 |
Crystal Structure Of The Mutant D70N Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With Sulfate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2009-06-23 Classification: LYASE Ligands: SO4 |
 |
Crystal Structure Of The Mutant D70N Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With Uridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-06-23 Classification: LYASE Ligands: U5P |
 |
Crystal Structure Of The Mutant D70N Of Orotidine 5'-Monophosphate Decarboxylase From Methanobacterium Thermoautotrophicum Complexed With 6-Azauridine 5'-Monophosphate
Organism: Methanothermobacter thermautotrophicus str. delta h
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2009-06-23 Classification: LYASE Ligands: UP6 |
 |
Crystal Structure Of The Orotidine 5'-Monophosphate Decarboxylase From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2009-06-23 Classification: LYASE |
 |
Crystal Structure Of The Orotidine 5'-Monophosphate Decarboxylase From Saccharomyces Cerevisiae Complexed With 6-Azauridine 5'-Monophosphate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2009-06-23 Classification: LYASE Ligands: UP6 |
 |
Crystal Structure Of The D91N Mutant Of The Orotidine 5'-Monophosphate Decarboxylase From Saccharomyces Cerevisiae
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-06-23 Classification: LYASE |
 |
Crystal Structure Of The D91N Mutant Of The Orotidine 5'-Monophosphate Decarboxylase From Saccharomyces Cerevisiae Complexed With 6-Azauridine 5'-Monophosphate
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2009-06-23 Classification: LYASE Ligands: UP6 |
 |
Crystal Structure Of D-Allulose 6-Phosphate 3-Epimerase From Escherichia Coli K12 Complexed With D-Glucitol 6-Phosphate And Magnesium
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2009-02-24 Classification: ISOMERASE Ligands: S6P, MG |
 |
Crystal Structure Of D-Allulose 6-Phosphate 3-Epimerase From Escherichia Coli K-12
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2008-08-26 Classification: ISOMERASE Ligands: MG, SO4 |

