Search Count: 7
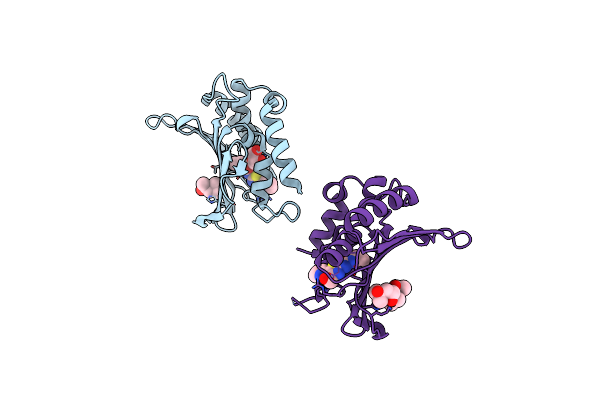 |
Crystal Structure Of The Atp Binding Domain Of S. Aureus Gyrb Complexed With A Ligand
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2015-11-25 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: 57U, MPD, GOL |
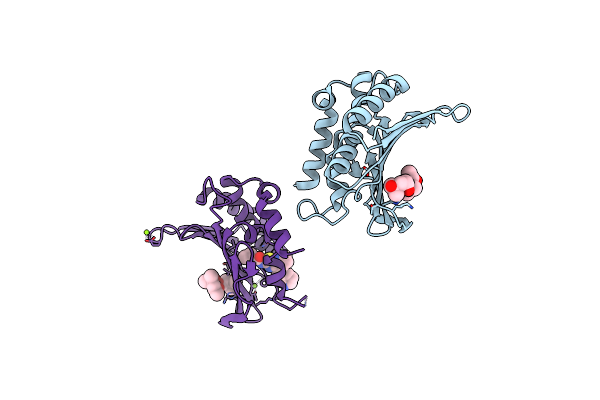 |
Crystal Structure Of The Atp Binding Domain Of S. Aureus Gyrb Complexed With A Ligand
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2015-11-25 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: MPD, 57V, MG |
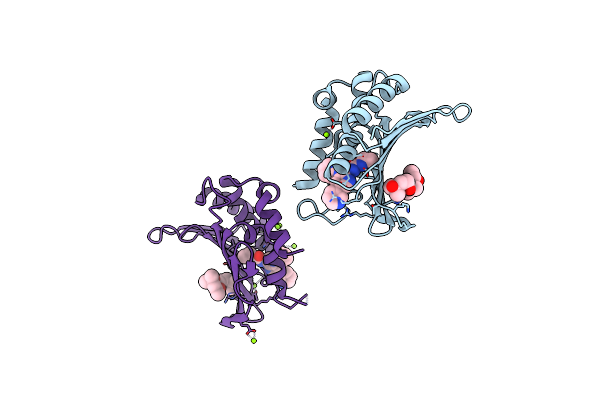 |
Crystal Structure Of The Atp Binding Domain Of S. Aureus Gyrb Complexed With A Ligand
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-11-25 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: MPD, 57W, MG |
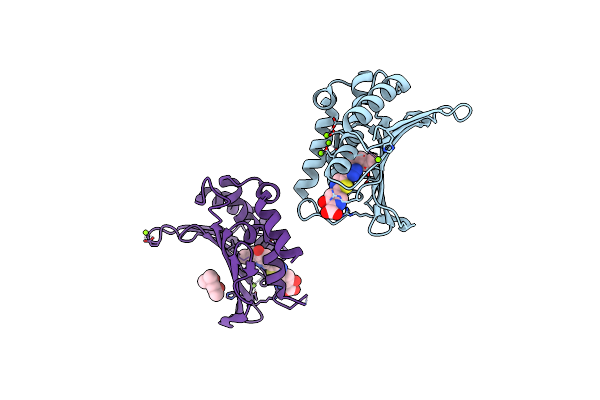 |
Crystal Structure Of The Atp Binding Domain Of S. Aureus Gyrb Complexed With A Ligand
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2015-11-18 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: MG, CL, 57Y, MPD |
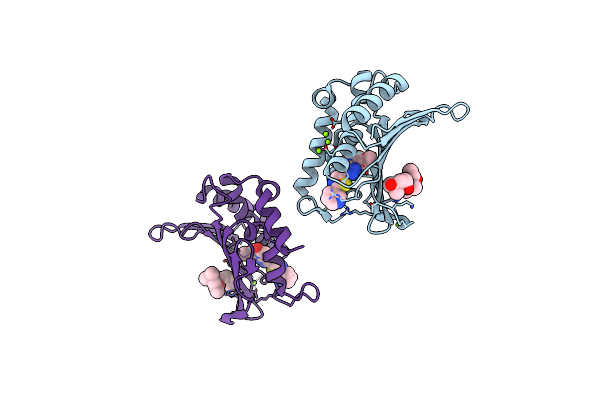 |
Crystal Structure Of The Atp Binding Domain Of S. Aureus Gyrb Complexed With A Ligand
Organism: Staphylococcus aureus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2015-11-11 Classification: ISOMERASE/ISOMERASE INHIBITOR Ligands: MPD, CL, MG, 57X |
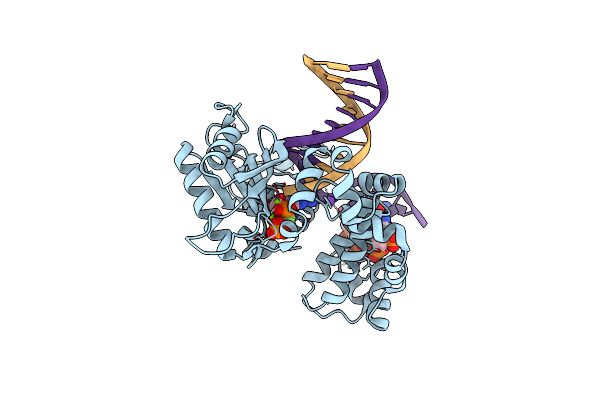 |
Ternary Complex Structure Of Dna Polymerase Beta With A Gapped Dna Substrate And A, B Damp(Cf2)Pp In The Active Site
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-03-21 Classification: TRANSFERASE/DNA Ligands: F3A, MG, NA, CL |
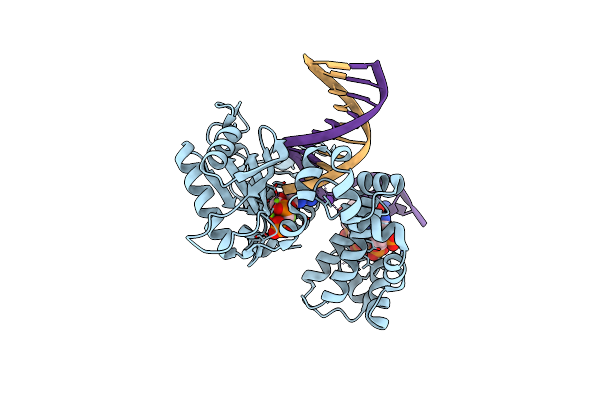 |
Ternary Complex Structure Of Dna Polymerase Beta With A Gapped Dna Substrate And A, B Damp(Cfh)Pp In The Active Site: Stereoselective Binding Of (S) Isomer
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2012-03-21 Classification: TRANSFERASE/DNA Ligands: FHA, MG, NA, CL |

