Search Count: 9
 |
Natively Purified Rubrerythrin 16-Mer From The Anaerobic Extremophile P. Furiosus
Organism: Pyrococcus furiosus
Method: ELECTRON MICROSCOPY Release Date: 2025-07-16 Classification: OXIDOREDUCTASE Ligands: FE |
 |
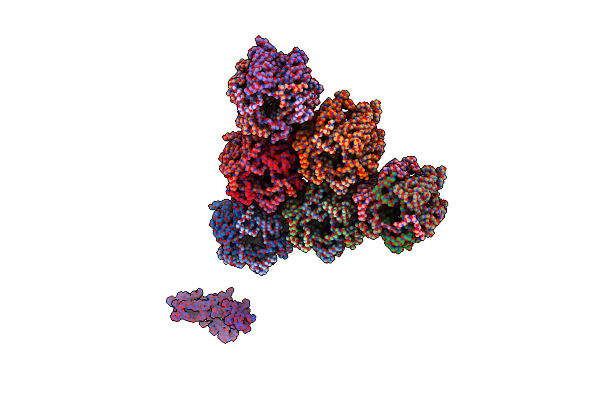
Emiliania Huxleyi Virus 201 (Ehv-201) Asymmetrical Unit Of Capsid Proteins Predicted By Alphafold2 Fitted Into The Cryo-Em Density Of Ehv-201 Virion Composite Map.
Organism: Emiliania huxleyi virus 201
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: VIRUS |
 |
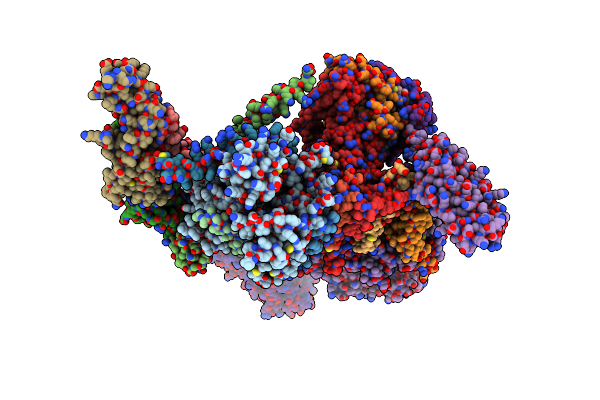
Emiliania Huxleyi Virus 201 (Ehv-201) Capsid Proteins Predicted By Alphafold2 Fitted Into A Cryo-Em Density Map Of The Ehv-201 Virion Capsid.
Organism: Emiliania huxleyi virus 201
Method: ELECTRON MICROSCOPY Release Date: 2023-12-20 Classification: VIRUS |
 |
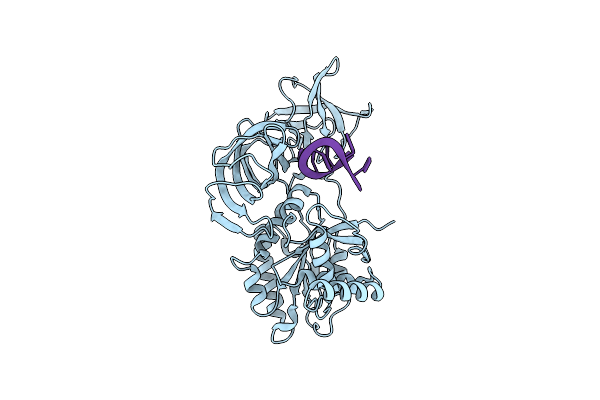
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: MEMBRANE PROTEIN |
 |
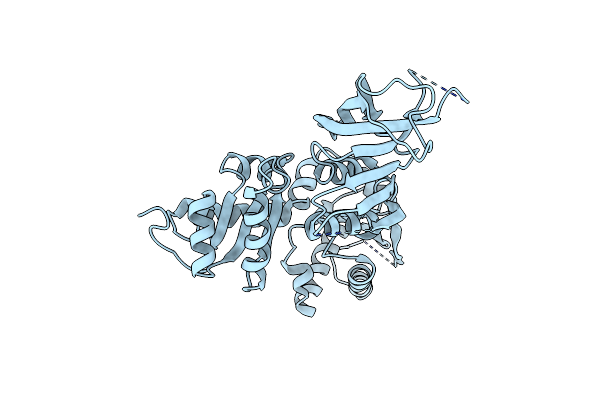
Cryoem Structure Of Extended Eef1A Bound To The Ribosome In The Classical Pre State
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-12-07 Classification: RIBOSOME |
 |
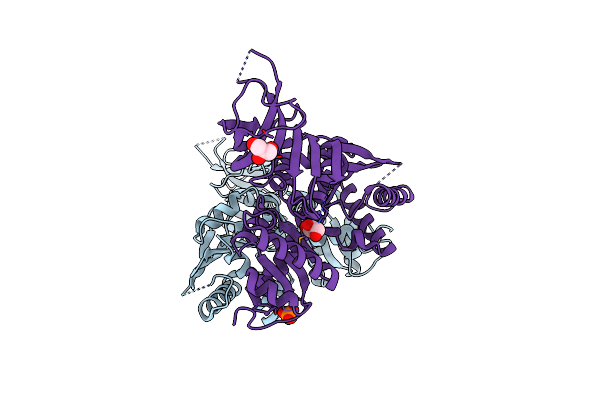
Structure Of S. Cerevisiae Dom34, A Translation Termination-Like Factor Involved In Rna Quality Control Pathways And Interacting With Hbs1 (Unlabeled Protein)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2008-01-22 Classification: CELL CYCLE |
 |
Structure Of S. Cerevisiae Dom34, A Translation Termination-Like Factor Involved In Rna Quality Control Pathways And Interacting With Hbs1 (Selenomet-Labeled Protein)
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.51 Å Release Date: 2008-01-22 Classification: CELL CYCLE Ligands: GOL, PO4 |
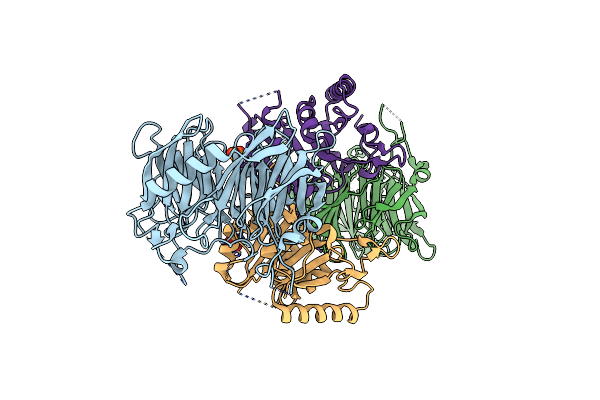 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2007-12-18 Classification: TRANSFERASE Ligands: PO4 |
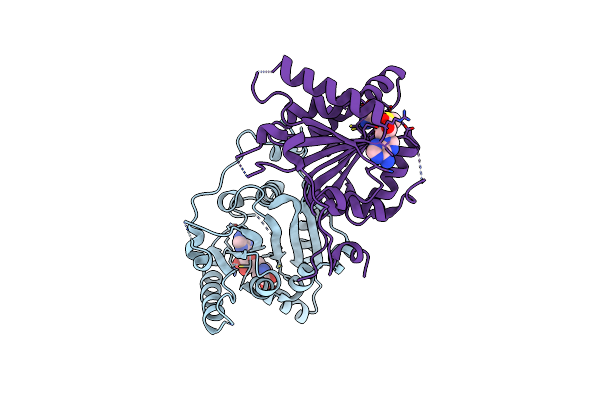 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2007-12-18 Classification: TRANSFERASE Ligands: SAM |

