Search Count: 170
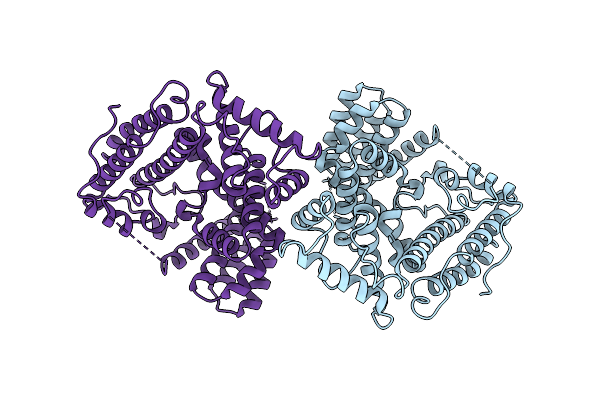 |
Organism: Puccinia graminis f. sp. tritici
Method: ELECTRON MICROSCOPY Release Date: 2025-09-24 Classification: ANTIFUNGAL PROTEIN |
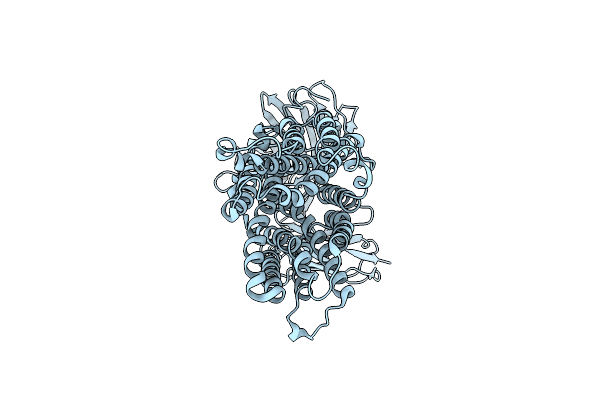 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: SPD |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: YHL |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2025-06-25 Classification: MEMBRANE PROTEIN Ligands: SPM |
 |
Organism: Hordeum vulgare, Blumeria graminis
Method: ELECTRON MICROSCOPY Release Date: 2025-02-12 Classification: ANTIFUNGAL PROTEIN |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2024-12-11 Classification: PLANT PROTEIN/HYDROLASE Ligands: APR, ATP |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Release Date: 2024-12-11 Classification: PLANT PROTEIN/HYDROLASE Ligands: APR, ATP |
 |
Organism: Oryza sativa japonica group
Method: ELECTRON MICROSCOPY Release Date: 2024-11-13 Classification: PLANT PROTEIN Ligands: A1D8A |
 |
Organism: Arabidopsis thaliana
Method: ELECTRON MICROSCOPY Resolution:3.34 Å Release Date: 2024-10-30 Classification: TRANSFERASE/PLANT PROTEIN Ligands: NAG |
 |
Organism: Solanum lycopersicum
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: PLANT PROTEIN Ligands: ADP, IHP |
 |
Organism: Solanum lycopersicum
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: PLANT PROTEIN Ligands: IHP, ADP |
 |
Organism: Solanum lycopersicum
Method: ELECTRON MICROSCOPY Release Date: 2024-05-22 Classification: PLANT PROTEIN Ligands: IHP, ADP |
 |
Organism: Phaseolus vulgaris, Fusarium phyllophilum
Method: X-RAY DIFFRACTION Resolution:1.94 Å Release Date: 2024-02-07 Classification: HYDROLASE |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2024-02-07 Classification: HYDROLASE Ligands: NAG |
 |
Organism: Arabidopsis thaliana
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2023-11-08 Classification: HYDROLASE Ligands: APR |
 |
Organism: Escherichia coli k-12, Escherichia coli
Method: ELECTRON MICROSCOPY Release Date: 2023-10-18 Classification: TRANSCRIPTION Ligands: PLP, ADP |
 |
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION Ligands: HOH |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Tritici Avrpm2(1)
Organism: Blumeria graminis f. sp. tritici
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |
 |
Crystal Structure Of Powdery Mildews Blumeria Graminis F. Sp. Hordei Avra22
Organism: Blumeria hordei
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2023-08-16 Classification: UNKNOWN FUNCTION |

