Search Count: 20
 |
Organism: Salasvirus phi29
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2024-01-17 Classification: DNA BINDING PROTEIN |
 |
Organism: Salasvirus phi29
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-01-17 Classification: DNA BINDING PROTEIN |
 |
Partial Structure Of Tyrosine Hydroxylase Lacking The First 35 Residues In Complex With Dopamine.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-22 Classification: OXIDOREDUCTASE Ligands: LDP, FE |
 |
Partial Structure Of Tyrosine Hydroxylase In Complex With Dopamine Showing The Catalytic Domain And An Alpha-Helix From The Regulatory Domain Involved In Dopamine Binding.
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-08 Classification: OXIDOREDUCTASE Ligands: LDP, FE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-12-01 Classification: OXIDOREDUCTASE Ligands: FE |
 |
Atomic Model Of The Em-Based Structure Of The Full-Length Tyrosine Hydroxylase In Complex With Dopamine (Residues 40-497) In Which The Regulatory Domain (Residues 40-165) Has Been Included Only With The Backbone Atoms
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: OXIDOREDUCTASE Ligands: FE, LDP |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-11-17 Classification: OXIDOREDUCTASE Ligands: FE |
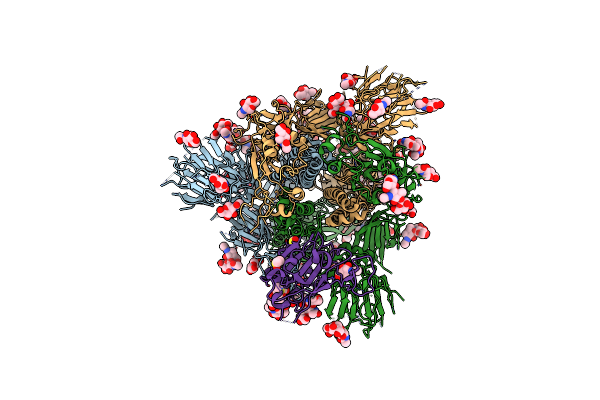 |
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: NAG, MAN, DMS |
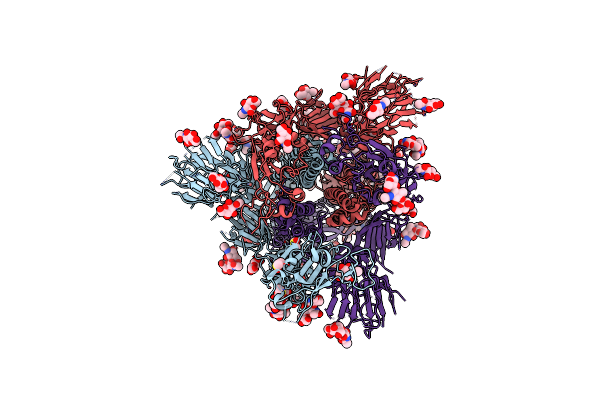 |
Sars-Cov-2 Spike In Prefusion State (Flexibility Analysis, 1-Up Closed Conformation)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: NAG, MAN, DMS |
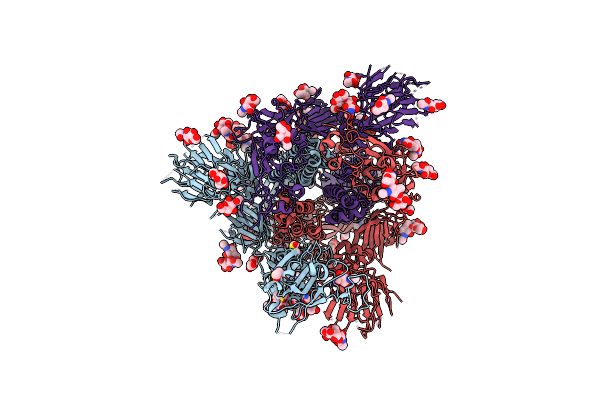 |
Sars-Cov-2 Spike In Prefusion State (Flexibility Analysis, 1-Up Open Conformation)
Organism: Severe acute respiratory syndrome coronavirus 2
Method: ELECTRON MICROSCOPY Release Date: 2020-07-29 Classification: VIRAL PROTEIN Ligands: NAG, MAN, DMS |
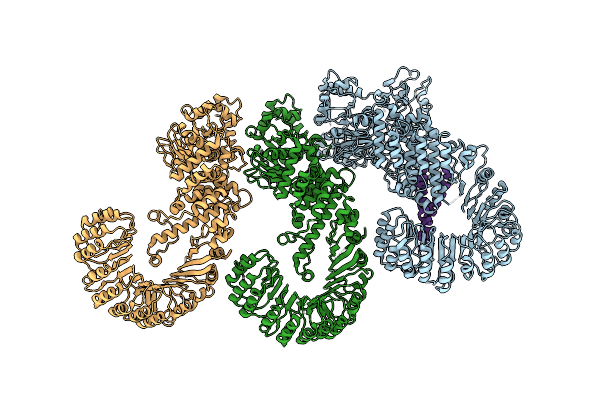 |
Organism: Mus musculus, Legionella pneumophila
Method: ELECTRON MICROSCOPY Release Date: 2017-11-15 Classification: IMMUNE SYSTEM |
 |
Organism: Homo sapiens, Sus scrofa
Method: ELECTRON MICROSCOPY Release Date: 2016-08-03 Classification: STRUCTURAL PROTEIN Ligands: GTP, MG, GDP, POU |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Resolution:8.50 Å Release Date: 2016-04-06 Classification: TRANSCRIPTION |
 |
Crystallographic Structure Of The Human Igg1 Alpha 2-6 Sialilated Fc-Fragment
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2014-11-12 Classification: IMMUNE SYSTEM |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2014-10-15 Classification: MEMBRANE PROTEIN Ligands: CA, SO4, GQ1 |
 |
Crystallographic Structure Of The Mouse Sign-R1 Crd Domain In Complex With Sialic Acid
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2014-10-15 Classification: IMMUNE SYSTEM Ligands: CA, SO4, SIA, CL |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.87 Å Release Date: 2014-01-15 Classification: IMMUNE SYSTEM Ligands: SO4, CA |
 |
Formation Of An Intricate Helical Bundle Dictates The Assembly Of The 26S Proteasome Lid
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2013-08-28 Classification: PROTEIN BINDING |
 |
Crystal Structure Of The Replication Initiator Protein Encoded On Plasmid Pmv158 (Repb), Trigonal Form, To 2.7 Ang Resolution
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2009-06-30 Classification: REPLICATION Ligands: MN, CL, MG |
 |
Crystal Structure Of The Replication Initiator Protein Encoded On Plasmid Pmv158 (Repb), Tetragonal Form, To 3.6 Ang Resolution
Organism: Streptococcus agalactiae
Method: X-RAY DIFFRACTION Resolution:3.60 Å Release Date: 2009-06-30 Classification: REPLICATION Ligands: MN |

