Search Count: 20
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-05-10 Classification: VIRAL PROTEIN |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN Ligands: CA, ZN |
 |
Organism: Rotavirus a
Method: ELECTRON MICROSCOPY Release Date: 2023-04-05 Classification: VIRAL PROTEIN |
 |
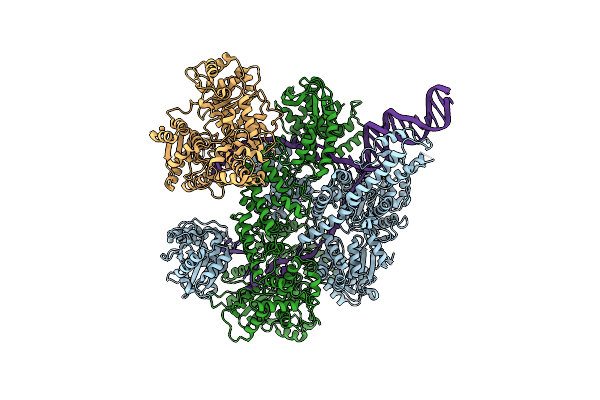
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-01-01 Classification: HYDROLASE |
 |
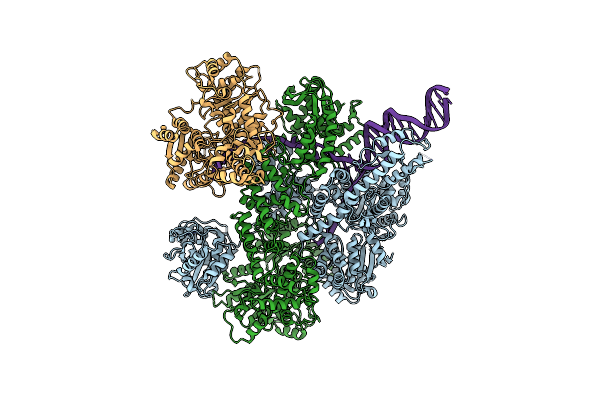
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-01-01 Classification: HYDROLASE |
 |
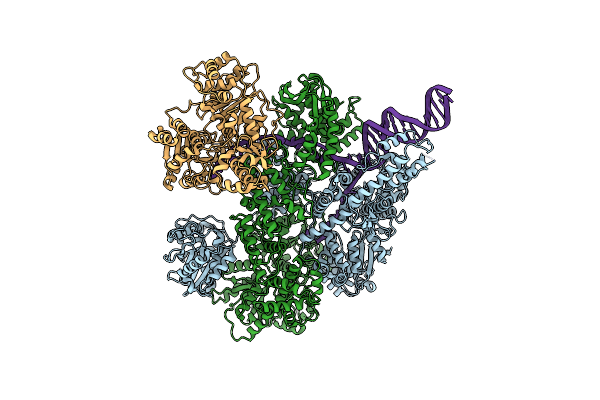
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-01-01 Classification: HYDROLASE |
 |
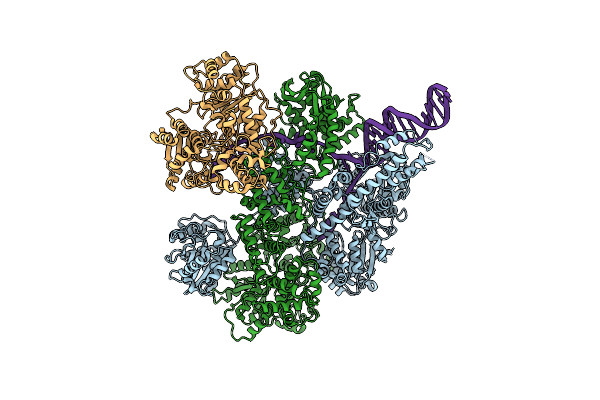
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-01-01 Classification: HYDROLASE |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-01-01 Classification: HYDROLASE |
 |
Organism: Escherichia coli, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2020-01-01 Classification: HYDROLASE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-12-25 Classification: CELL CYCLE |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2019-12-25 Classification: CELL CYCLE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-10-17 Classification: NUCLEAR PROTEIN Ligands: ADP, BEF, MG, ZN |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2018-10-17 Classification: NUCLEAR PROTEIN Ligands: ADP, BEF, MG, ZN |
 |
Organism: Escherichia coli, Enterobacteria phage lambda
Method: ELECTRON MICROSCOPY Release Date: 2017-01-11 Classification: DNA BINDING PROTEIN |
 |
Cryo-Em Structure Of Recbcd+Dna Complex Revealing Activated Nuclease Domain
Organism: Escherichia coli (strain k12), Endothia gyrosa
Method: ELECTRON MICROSCOPY Release Date: 2016-10-05 Classification: HYDROLASE Ligands: ANP, MG |
 |
Structural Basis For Dna Strand Separation By A Hexameric Replicative Helicase
Organism: Bovine papillomavirus
Method: ELECTRON MICROSCOPY Resolution:19.00 Å Release Date: 2015-08-26 Classification: HYDROLASE Ligands: PO4, MG |
 |
Structure Of Bacteriophage Spp1 Head-To-Tail Interface Filled With Dna And Tape Measure Protein
Organism: Bacillus phage spp1
Method: ELECTRON MICROSCOPY Resolution:7.60 Å Release Date: 2015-06-03 Classification: VIRAL PROTEIN |
 |
Structure Of Bacteriophage Spp1 Head-To-Tail Interface Without Dna And Tape Measure Protein
Organism: Bacillus phage spp1
Method: ELECTRON MICROSCOPY Resolution:7.20 Å Release Date: 2015-06-03 Classification: VIRAL PROTEIN |
 |
Cryo-Em Map Of A Yeast Minimal Preinitiation Complex Interacting With The Mediator Head Module
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2013-03-06 Classification: TRANSFERASE |
 |
Cryo-Em Map Of A Yeast Minimal Preinitiation Complex Interacting With The Mediator Head Module
Organism: Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2013-03-06 Classification: TRANSCRIPTION |

