Search Count: 20
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-03 Classification: Blood Clotting, Hydrolase Ligands: OQ6 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: OQI, CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, hydrolase Ligands: OQO, CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: OR0, CIT |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.59 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: CIT, ORF |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: CIT, ORU |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: CIT, OSX |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: CIT, OT7 |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: CIT, OTL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.73 Å Release Date: 2022-08-03 Classification: BLOOD CLOTTING, Hydrolase Ligands: CIT, OTX |
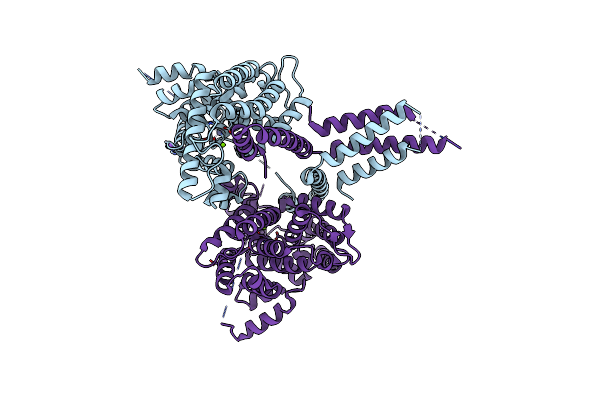 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2015-03-11 Classification: HYDROLASE Ligands: ZN, NA, MG, IOD |
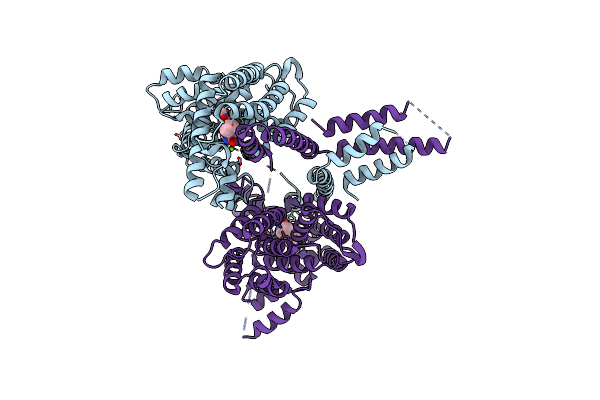 |
Crystal Structure Of Crosslink Stabilized Long-Form Pde4B In Complex With (R)-(-)-Rolipram
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:3.22 Å Release Date: 2015-03-11 Classification: hydrolase/hydrolase inhibitor Ligands: ZN, IOD, ROL, MG |
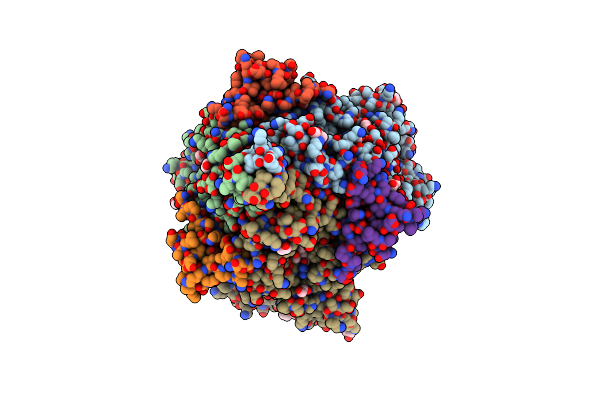 |
Complex Crystal Structure Of Hydroxylamine Oxidoreductase And Ne1300 From Nitrosomonas Europaea
Organism: Nitrosomonas europaea
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2013-09-04 Classification: OXIDOREDUCTASE Ligands: HEC, ISW, PEG, PGE, EDO, P6G, PG4, NO3 |
 |
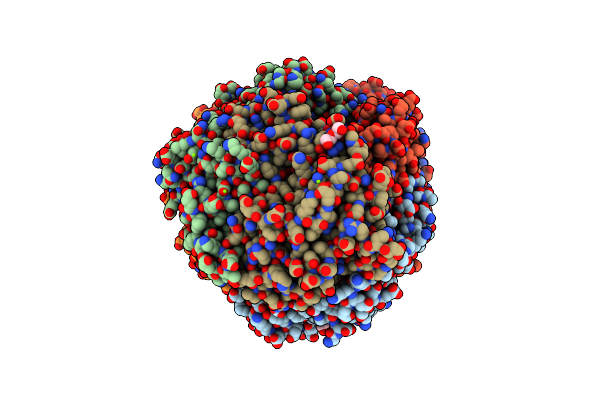
Structural Analysis Of A Ni(Iii)-Methyl Species In Methyl-Coenzyme M Reductase From Methanothermobacter Marburgensis
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2011-04-13 Classification: TRANSFERASE Ligands: MG, F43, COM, TXZ, TP7, 06C, K, EDO |
 |
Structural Insight Into Methyl-Coenzyme M Reductase Chemistry Using Coenzyme B Analogues
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2010-09-15 Classification: TRANSFERASE Ligands: MG, F43, TP7, COM, ACT, PEG, ZN, EDO |
 |
Structural Insight Into Methyl-Coenzyme M Reductase Chemistry Using Coenzyme B Analogues
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.30 Å Release Date: 2010-09-15 Classification: TRANSFERASE Ligands: MG, F43, TP7, COM, TPZ, EDO, ZN, PEG |
 |
Structural Insight Into Methyl-Coenzyme M Reductase Chemistry Using Coenzyme B Analogues
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2010-09-15 Classification: TRANSFERASE Ligands: MG, F43, TP7, COM, TXZ, ACT, EDO, PEG, ZN |
 |
Structural Insight Into Methyl-Coenzyme M Reductase Chemistry Using Coenzyme B Analogues
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2010-09-15 Classification: TRANSFERASE Ligands: MG, F43, TP7, COM, XP8, ACT, EDO, ZN |
 |
Structural Insight Into Methyl-Coenzyme M Reductase Chemistry Using Coenzyme B Analogues
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2010-09-15 Classification: TRANSFERASE Ligands: MG, F43, TP7, COM, XP9, ACT, EDO, ZN, PEG |
 |
Structural Insight Into Methyl-Coenzyme M Reductase Chemistry Using Coenzyme B Analogues
Organism: Methanothermobacter marburgensis
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2010-09-15 Classification: TRANSFERASE Ligands: MG, TP7, F43, COM, SHT, ACT, EDO, ZN, PEG |

