Search Count: 44
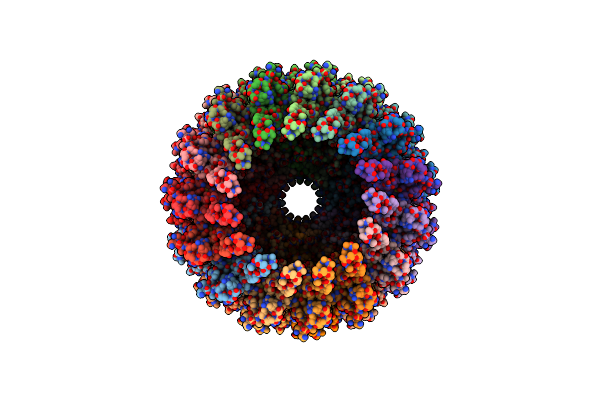 |
Organism: Deinococcus radiodurans r1 = atcc 13939 = dsm 20539
Method: ELECTRON MICROSCOPY Release Date: 2024-04-10 Classification: MEMBRANE PROTEIN |
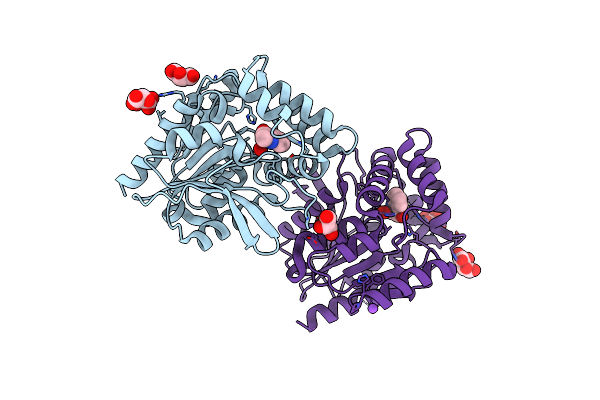 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) S101A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Hyperoxyc Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: VFH, OXY, TAR, GOL, NA |
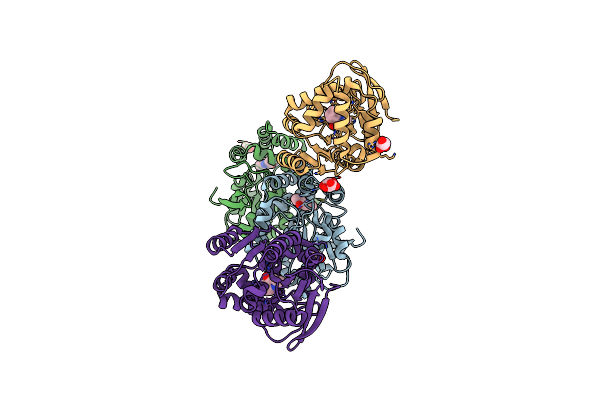 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) S101A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Normoxyc Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: VFH, GOL, SRT |
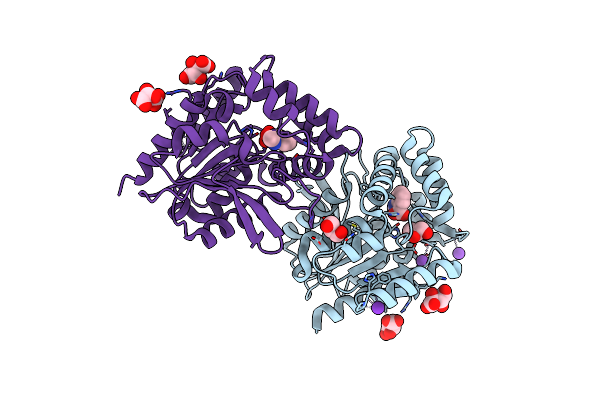 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) H251A Variant Complexed With N-Acetylanthranilate As Result Of In Crystallo Turnover Of Its Natural Substrate 1-H-3-Hydroxy-4- Oxoquinaldine Under Hyperoxic Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2024-01-17 Classification: OXIDOREDUCTASE Ligands: ZZ8, SRT, GOL, K |
 |
Room Temperature Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Under Xenon Pressure (30 Bar)
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.90 Å Release Date: 2023-07-05 Classification: OXIDOREDUCTASE Ligands: XE, TAR |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Catalytically Inactive H251A Variant Complexed With 2-Methyl-Quinolin-4(1H)-One Under Normoxic Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-06-01 Classification: OXIDOREDUCTASE Ligands: VFH, K, GOL, SRT, NA |
 |
Crystal Structure Of The Cofactor-Devoid 1-H-3-Hydroxy-4- Oxoquinaldine 2,4-Dioxygenase (Hod) Catalytically Inactive H251A Variant Complexed With 2-Methyl- Quinolin-4(1H)-One Under Hyperoxic Conditions
Organism: Paenarthrobacter nitroguajacolicus
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-06-01 Classification: OXIDOREDUCTASE Ligands: VFH, K, TAR, GOL |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2022-03-09 Classification: SIGNALING PROTEIN Ligands: NAG |
 |
Crystal Structure Of The Ferric Enterobactin Receptor (Pfea) From Pseudomonas Aeruginosa In Complex With Tcv-L6
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.66 Å Release Date: 2021-08-04 Classification: MEMBRANE PROTEIN Ligands: V82, FE |
 |
Crystal Structure Of The Ferric Enterobactin Receptor (Pfea) In Complex With Bcv-L5
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.04 Å Release Date: 2021-01-20 Classification: MEMBRANE PROTEIN Ligands: OWT, EDO, FE |
 |
Crystal Structure Of The Ferric Enterobactin Receptor (Pfea) In Complex With Bcv-L6
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:3.03 Å Release Date: 2021-01-20 Classification: MEMBRANE PROTEIN Ligands: EDO, Q5N, FE |
 |
Crystal Structure Of The Ferric Enterobactin Receptor (Pfea) In Complex With Tcv_L5
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.72 Å Release Date: 2020-07-29 Classification: MEMBRANE PROTEIN Ligands: FE, QDQ |
 |
Crystal Structure Of The Ferric Enterobactin Receptor (Pfea) In Complex With Bcv
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2020-07-29 Classification: MEMBRANE PROTEIN Ligands: Q62, EDO, FE |
 |
Crystal Structure Of The Ferric Enterobactin Receptor Mutant R480A From Pseudomonas Aeruginosa (Pfea) In Complex With Enterobactin
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:3.11 Å Release Date: 2019-04-10 Classification: MEMBRANE PROTEIN Ligands: FE, EB4 |
 |
Crystal Structure Of The Ferric Enterobactin Receptor Mutant (Q482A) From Pseudomonas Aeruginosa (Pfea) In Complex With Enterobactin
Organism: Pseudomonas aeruginosa pao1
Method: X-RAY DIFFRACTION Resolution:2.96 Å Release Date: 2019-01-16 Classification: MEMBRANE PROTEIN Ligands: FE, EB4 |
 |
Crystal Structure Of The Ferric Enterobactin Receptor From Pseudomonas Aeruginosa (Pfea) In Complex With Enterobactin
Organism: Pseudomonas aeruginosa (strain atcc 15692 / dsm 22644 / cip 104116 / jcm 14847 / lmg 12228 / 1c / prs 101 / pao1)
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-01-16 Classification: MEMBRANE PROTEIN Ligands: FE, EB4, LP5 |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2018-10-31 Classification: TRANSPORT PROTEIN Ligands: SO4, BOG, C8E |
 |
Crystal Structure Of Baua, The Ferric Preacinetobactin Receptor From Acinetobacter Baumannii In Complex With Fe3+-Preacinetobactin-Acinetobactin
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2018-10-10 Classification: MEMBRANE PROTEIN Ligands: FE, FV8, OPV, OPZ, C8E, EDO |
 |
Crystal Structure Of Baua, The Ferric Preacinetobactin Receptor From Acinetobacter Baumannii
Organism: Acinetobacter baumannii (strain atcc 19606 / dsm 30007 / cip 70.34 / jcm 6841 / nbrc 109757 / ncimb 12457 / nctc 12156 / 81)
Method: X-RAY DIFFRACTION Resolution:2.54 Å Release Date: 2018-10-10 Classification: MEMBRANE PROTEIN Ligands: EDO, C8E |
 |
Crystal Structure Of Baua, The Ferric Preacinetobactin Receptor From Acinetobacter Baumannii
Organism: Acinetobacter baumannii
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-10-10 Classification: MEMBRANE PROTEIN Ligands: EDO, C8E, PO4 |

