Search Count: 50
 |
Organism: Caenorhabditis elegans
Method: ELECTRON MICROSCOPY Release Date: 2024-06-26 Classification: HYDROLASE |
 |
Cryo-Em Structure Of A 1033 Scaffold Base Dna Origami Nanostructure V4 And Tba
Organism: Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2024-04-24 Classification: DNA |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-06-14 Classification: LIGASE Ligands: ZN |
 |
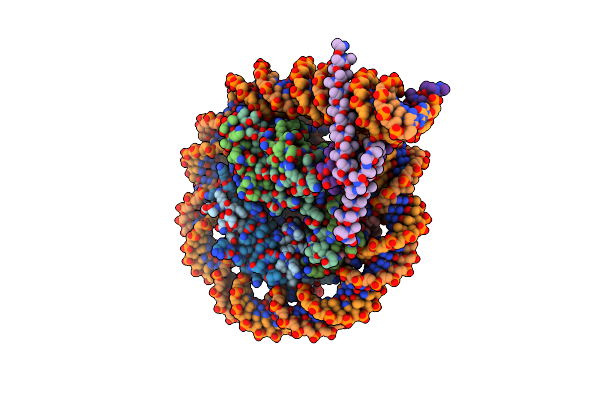 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl-6.2 (Dna Conformation 1)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
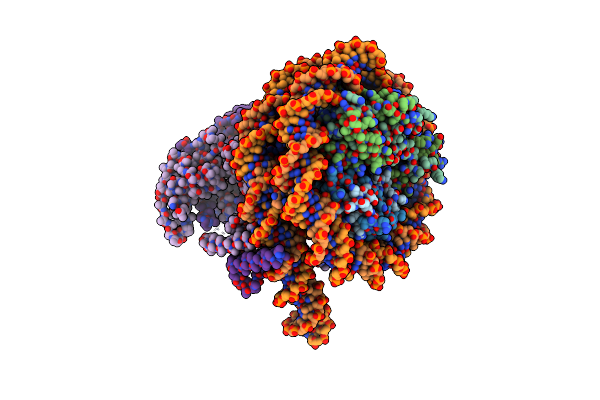 |
Cryo-Em Structure Of Clock-Bmal1 Bound To A Nucleosomal E-Box At Position Shl+5.8 (Composite Map)
Organism: Homo sapiens, Mus musculus, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
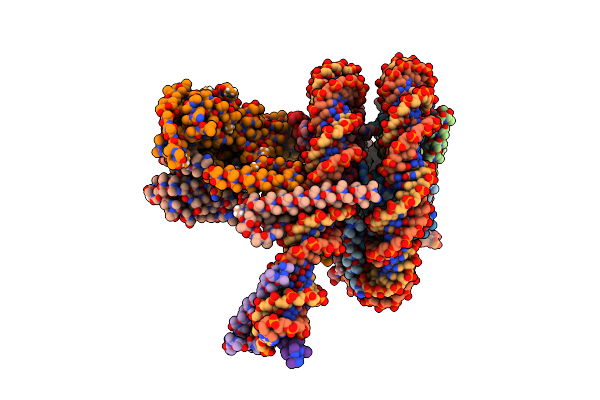 |
Cryo-Em Structure Of Clock-Bmal1 Bound To The Native Por Enhancer Nucleosome (Map 2, Additional 3D Classification And Flexible Refinement)
Organism: Homo sapiens, Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: GENE REGULATION |
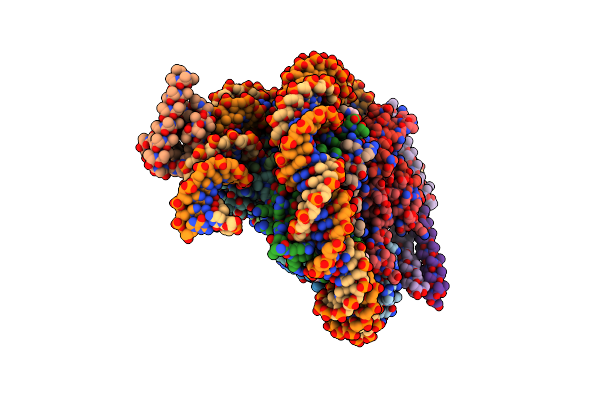 |
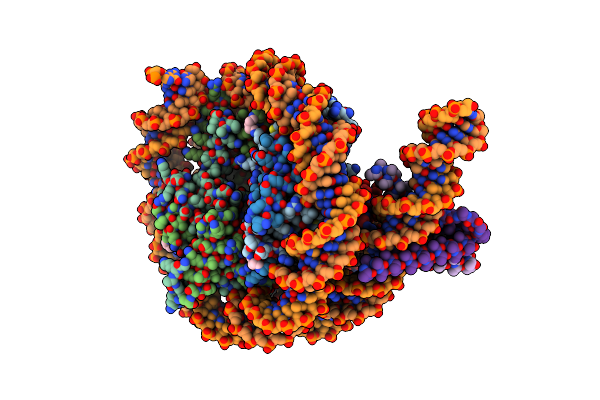
Organism: Homo sapiens, Aequorea victoria, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
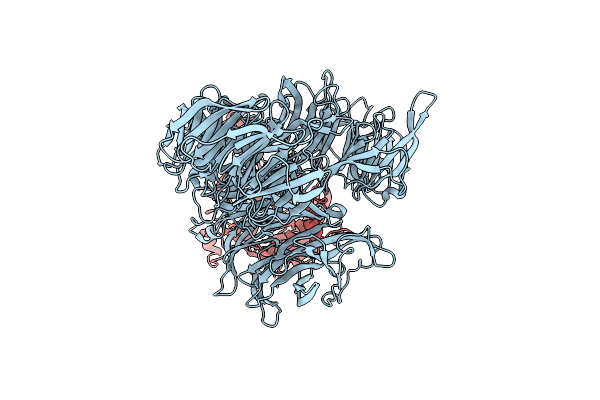
Organism: Homo sapiens, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2023-05-24 Classification: TRANSCRIPTION Ligands: PTD |
 |
 |
 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: PROTEIN TRANSPORT |
 |
Organism: Mus musculus
Method: ELECTRON MICROSCOPY Release Date: 2022-04-06 Classification: PROTEIN TRANSPORT Ligands: GTP |
 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: HYDROLASE Ligands: PO4 |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-08-04 Classification: HYDROLASE |
 |
Organism: Saccharomyces cerevisiae s288c, Homo sapiens, Escherichia coli k-12
Method: ELECTRON MICROSCOPY Release Date: 2021-07-21 Classification: PROTEIN BINDING |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2021-02-03 Classification: RIBOSOME Ligands: MG, ZN |
 |
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-09-23 Classification: IMMUNE SYSTEM Ligands: PTD, ZN |

