Search Count: 32
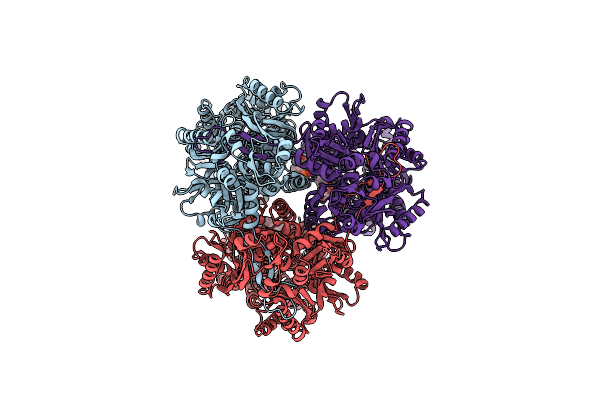 |
Single Particle Cryo-Em Co-Structure Of Klebsiella Pneumoniae Acrb With The Bdm91288 Efflux Pump Inhibitor At 2.97 Angstrom Resolution
Organism: Klebsiella pneumoniae subsp. pneumoniae dsm 30104 = jcm 1662 = nbrc 14940
Method: ELECTRON MICROSCOPY Release Date: 2024-01-17 Classification: TRANSPORT PROTEIN Ligands: WQW, WR6, PG8 |
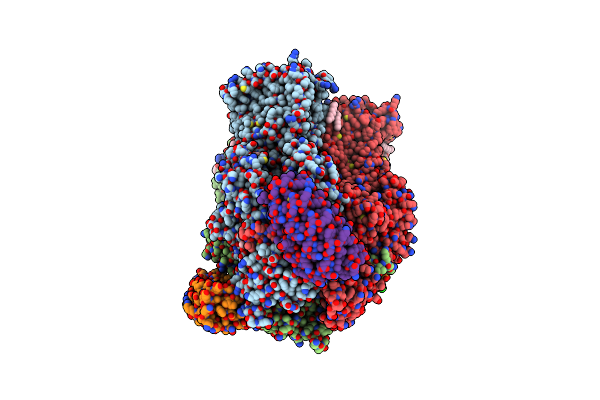 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2021-12-29 Classification: TRANSPORT PROTEIN Ligands: LMT, 1K8, EDO, OCT, C14, GOL, D10, DDR, DDQ, SO4, LPX |
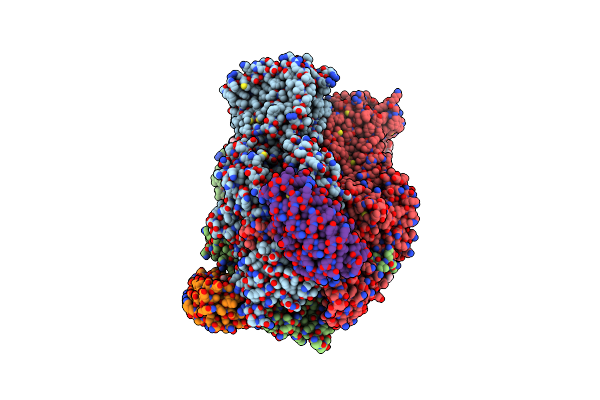 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2021-12-29 Classification: TRANSPORT PROTEIN Ligands: LMT, EDO, 1KE, D12, SO4, GOL, LPX, DDQ, C14 |
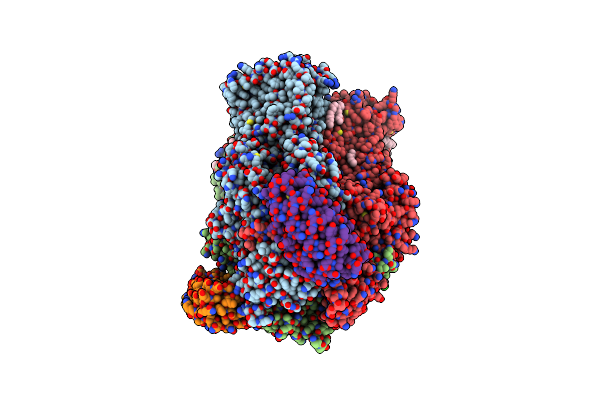 |
Organism: Escherichia coli, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.45 Å Release Date: 2021-12-29 Classification: TRANSPORT PROTEIN Ligands: LMT, C14, DDQ, GOL, 1K8, D10, EDO, SO4, OCT, LPX |
 |
Organism: Stenotrophomonas maltophilia
Method: X-RAY DIFFRACTION Resolution:1.50 Å Release Date: 2021-10-06 Classification: ANTIMICROBIAL PROTEIN Ligands: ZN, RBW, SO4 |
 |
Solution Nmr Structure Of De Novo Designed Rossmann 3X3 Fold Protein R3X3_Bp3, Northeast Structural Genomics Consortium (Nesg) Target Or689
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2021-05-19 Classification: DE NOVO PROTEIN |
 |
Structure Of The Sars-Cov-2 S 6P Trimer In Complex With The Ace2 Protein Decoy, Ctc-445.2 (State 4)
Organism: Severe acute respiratory syndrome coronavirus 2, Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2020-11-11 Classification: ANTIVIRAL PROTEIN Ligands: NAG |
 |
Solution Nmr Structure Of De Novo Designed Rossmann 2X3 Fold Protein R2X3_168, Northeast Structural Genomics Consortium (Nesg) Target Or386
Organism: Synthetic construct
Method: SOLUTION NMR Release Date: 2020-08-05 Classification: DE NOVO PROTEIN |
 |
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE |
 |
Alcohol Dehydrogenase From Candida Magnoliae Dsmz 70638 (Adha): Complex With Nadp+
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE Ligands: NAP, IPA, GOL |
 |
Alcohol Dehydrogenase From Candida Magnoliae Dsmz 70638 (Adha): Thermostable 10Fold Mutant
Organism: Starmerella magnoliae
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-04-08 Classification: OXIDOREDUCTASE |
 |
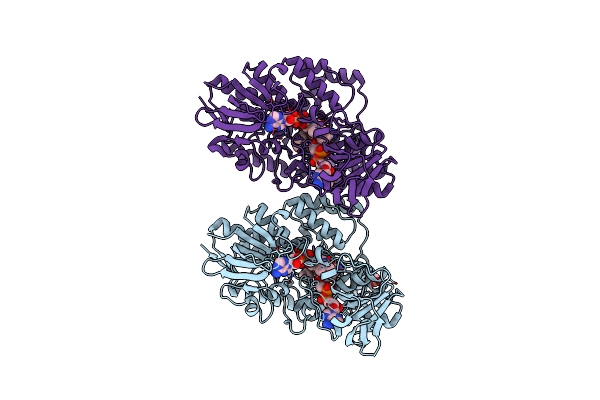
Structure Of A De Novo Designed Interleukin-2/Interleukin-15 Mimetic Complex With Il-2Rb And Il-2Rg
Organism: Synthetic construct, Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.52 Å Release Date: 2019-01-16 Classification: BIOSYNTHETIC PROTEIN/Protein Binding Ligands: NAG |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2019-01-16 Classification: BIOSYNTHETIC PROTEIN |
 |
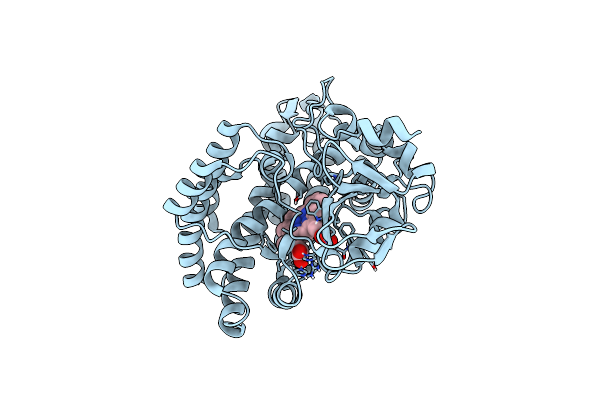
Thermocrispum Municipale Cyclohexanone Monooxygenase Bound To Hexanoic Acid
Organism: Thermocrispum municipale
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2018-12-12 Classification: FLAVOPROTEIN Ligands: FAD, NAP, 6NA, ACT |
 |
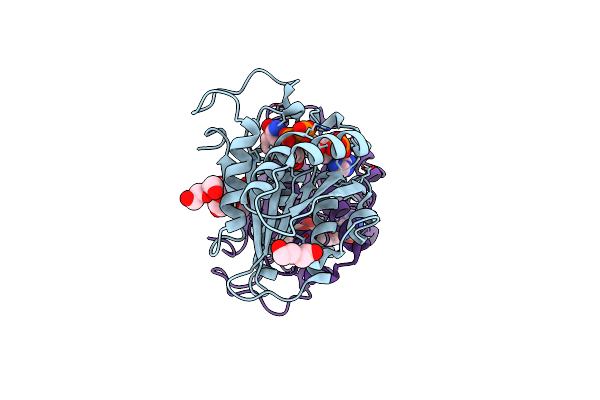
Organism: Tepidiphilus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-05-30 Classification: OXIDOREDUCTASE Ligands: HEM |
 |
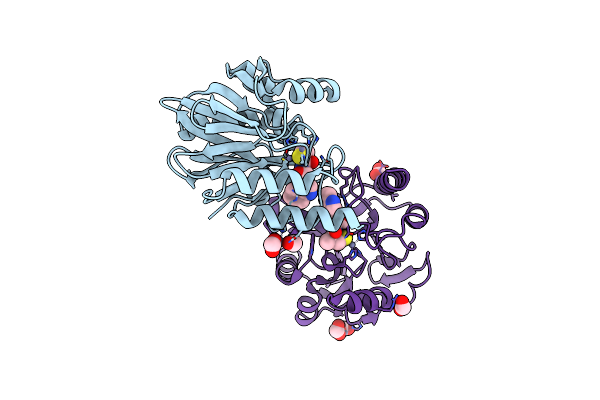
Organism: Candida magnoliae
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-12-20 Classification: OXIDOREDUCTASE Ligands: NAP, PEG |
 |
Vim-2 Metallo-Beta-Lactamase In Complex With ((S)-3-Mercapto-2-Methylpropanoyl)-D-Tryptophan (Compound 3)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.20 Å Release Date: 2017-05-17 Classification: HYDROLASE Ligands: ZN, R38, FMT |
 |
Vim-2 Metallo-Beta-Lactamase In Complex With ((S)-3-Mercapto-2-Methylpropanoyl)-L-Tryptophan (Compound 4)
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.16 Å Release Date: 2017-05-17 Classification: HYDROLASE Ligands: ZN, R59, FMT, BEZ |
 |
Mono-Zinc Vim-5 Metallo-Beta-Lactamase In Complex With (1-Chloro-4-Hydroxyisoquinoline-3-Carbonyl)-L-Tryptophan (Compound 2)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2017-05-17 Classification: HYDROLASE Ligands: ZN, 8NN |
 |
Di-Zinc Vim-5 Metallo-Beta-Lactamase In Complex With (1-Chloro-4-Hydroxyisoquinoline-3-Carbonyl)-D-Tryptophan (Compound 1)
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2017-05-17 Classification: HYDROLASE Ligands: ZN, 93W, GOL |

