Search Count: 58
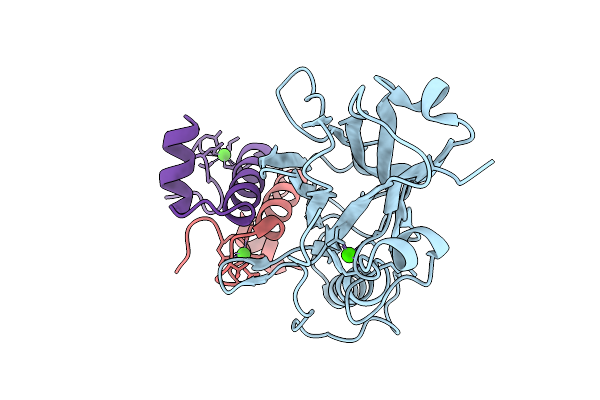 |
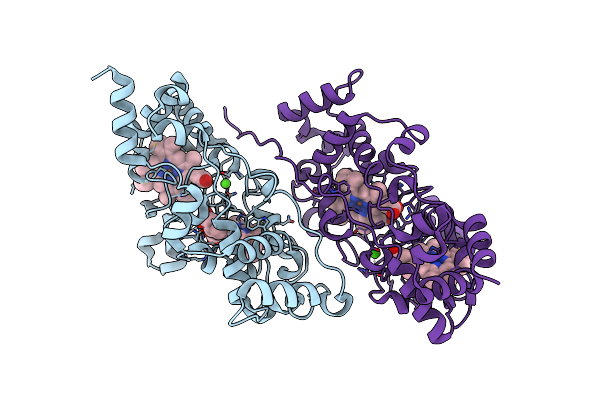
Ruminococcus Flavefaciens Coh-Doc Complex Between A Group 2 Dockerin And The Cohesin From Cell Surface Attached Scaffoldin Scae
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Release Date: 2025-10-08 Classification: PROTEIN BINDING Ligands: CA |
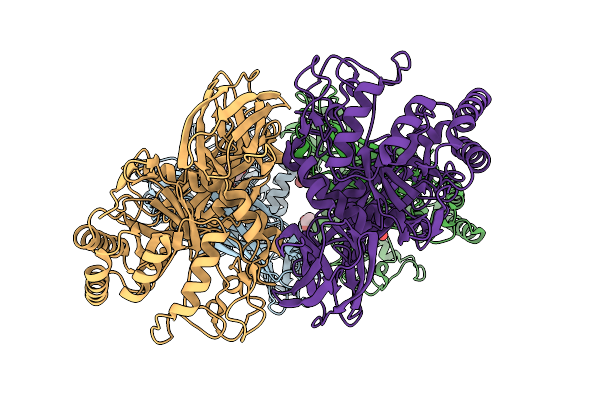 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum, Wild Type Semet Derivative (Lagh157)
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Release Date: 2025-07-16 Classification: HYDROLASE Ligands: ACT, GOL, MLA |
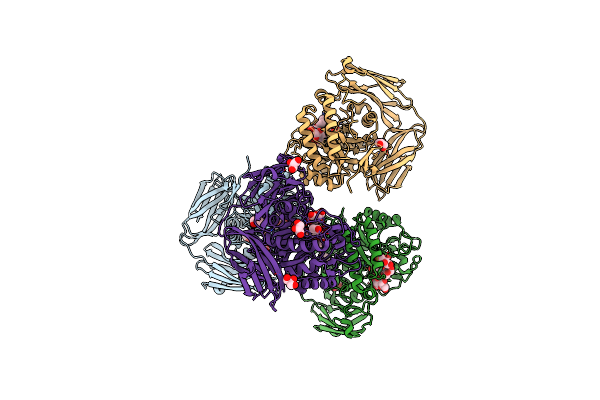 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) E224A Mutant In Complex With Laminaritriose And Glucose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.32 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: EDO, BGC, GOL, PGE |
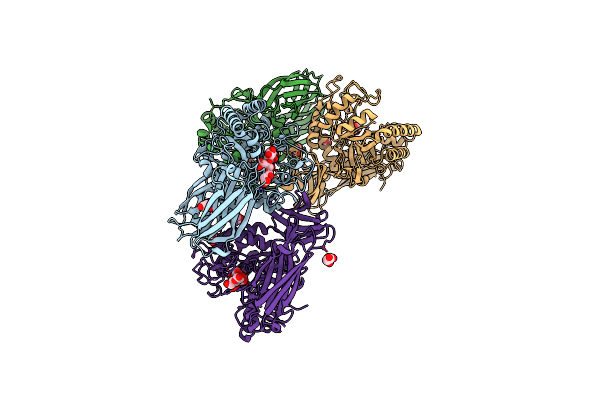 |
Glycoside Hydrolase Family 157 From Labilibaculum Antarcticum (Lagh157) In Complex With Laminaribiose
Organism: Labilibaculum antarcticum
Method: X-RAY DIFFRACTION Resolution:2.71 Å Release Date: 2025-05-21 Classification: HYDROLASE Ligands: MLA, GOL, BGC, PG4 |
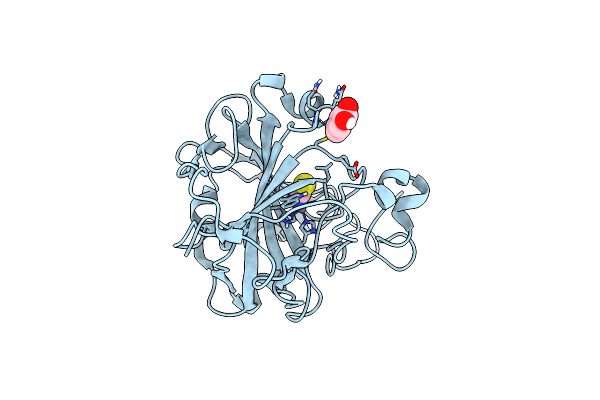 |
Structure Of Zinc(Ii) Double Mutant Human Carbonic Anhydrase Ii Bound To Thiocyanate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2024-02-07 Classification: METAL BINDING PROTEIN Ligands: HGB, SCN, ZN |
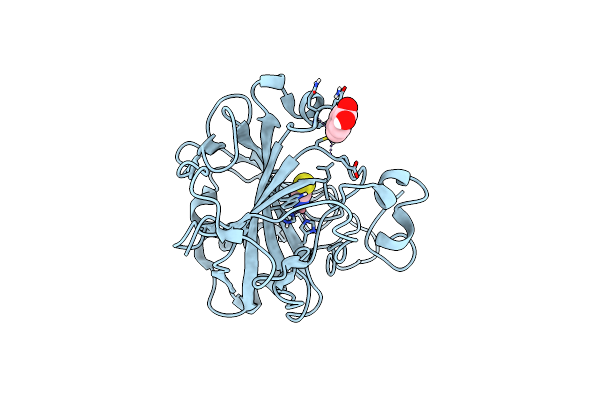 |
Structure Of Cobalt(Ii) Substituted Double Mutant Human Carbonic Anhydrase Ii Bound To Thiocyanate
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2024-02-07 Classification: METAL BINDING PROTEIN Ligands: HGB, CO, SCN |
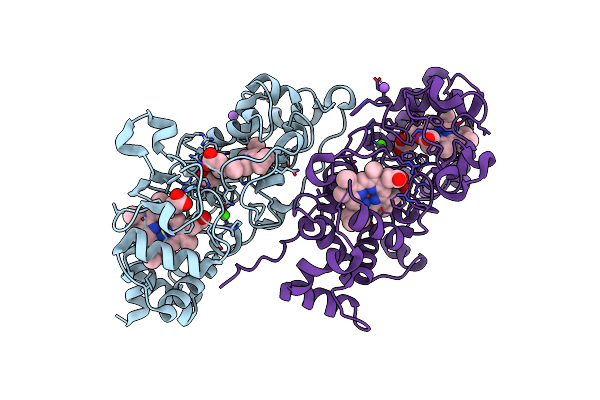 |
Mixed-Valence, Active Form, Of Cytochrome C Peroxidase From Obligate Human Pathogenic Bacterium Neisseria Gonorrhoeae At 1.4 Angstrom Resolution
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2023-04-12 Classification: ELECTRON TRANSPORT Ligands: HEC, CA, NA |
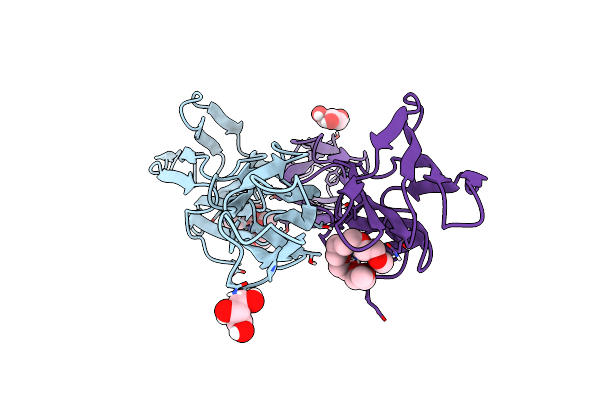 |
Crystal Structure Of The Dimeric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2023-02-22 Classification: HYDROLASE Ligands: PG4, GOL, PG6, CL |
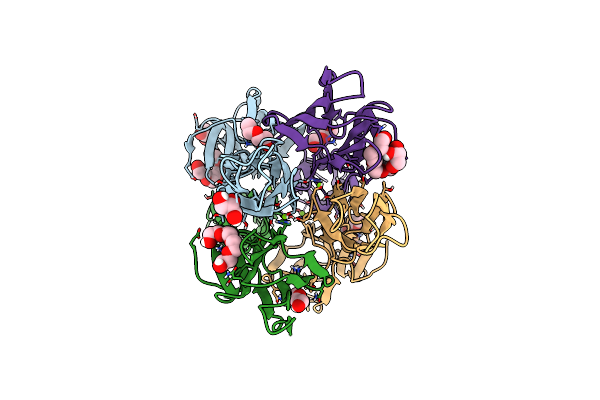 |
Crystal Structure Of The Tetrameric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2023-02-15 Classification: HYDROLASE Ligands: PG4, PEG, GOL, CL, MG |
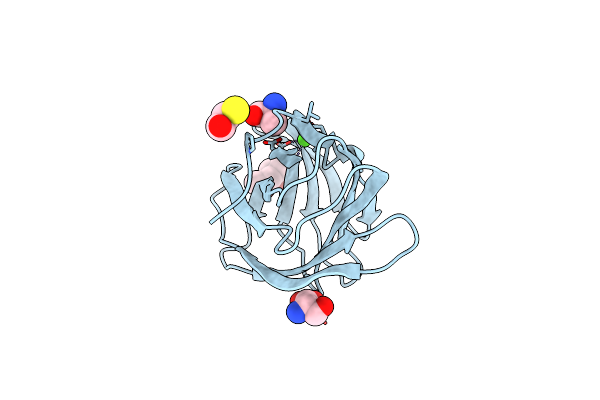 |
Organism: Bacteroides thetaiotaomicron vpi-5482
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2023-02-08 Classification: SUGAR BINDING PROTEIN Ligands: MPD, SER, BME, ALA, CA, CL |
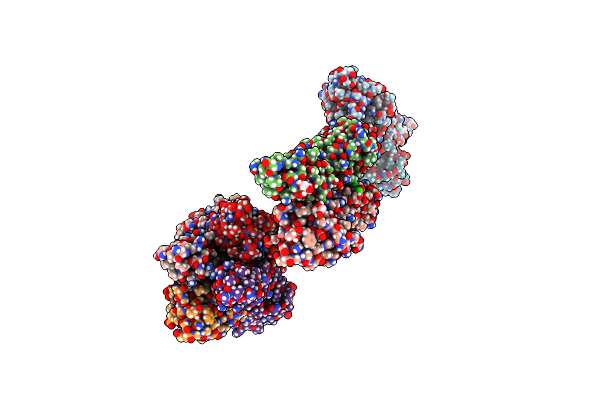 |
Crystal Structure Of The Semet Octameric C-Terminal Big_2-Cbm56 Domains From Paenibacillus Illinoisensis (Bacillus Circulans Iam1165) Beta-1,3-Glucanase H
Organism: Paenibacillus illinoisensis
Method: X-RAY DIFFRACTION Resolution:2.16 Å Release Date: 2023-02-01 Classification: HYDROLASE Ligands: GOL, CL |
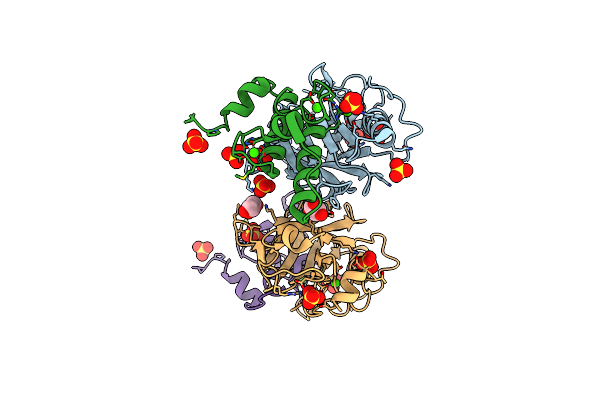 |
Ruminococcus Flavefaciens Cohesin-Dockerin Structure: Dockerin From Scah Adaptor Scaffoldin In Complex With The Cohesin From Scae Anchoring Scaffoldin
Organism: Ruminococcus flavefaciens fd-1
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2022-11-02 Classification: PROTEIN BINDING Ligands: CA, GOL, SO4 |
 |
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2022-01-26 Classification: APOPTOSIS Ligands: HEM, SO4 |
 |
Organism: Bacteroides ovatus (strain atcc 8483 / dsm 1896 / jcm 5824 / nctc 11153)
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2021-12-08 Classification: SUGAR BINDING PROTEIN Ligands: HED, BME, SO4, GOL, AZI |
 |
Structure Of Sgbp Bo2743 From Bacteroides Ovatus In Complex With Mixed-Linked Gluco-Nonasaccharide
Organism: Bacteroides ovatus atcc 8483
Method: X-RAY DIFFRACTION Resolution:1.43 Å Release Date: 2021-11-10 Classification: SUGAR BINDING PROTEIN Ligands: MG, GOL, PGE, NA, AZI, TAM |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.25 Å Release Date: 2021-04-21 Classification: SUGAR BINDING PROTEIN Ligands: GOL, CL |
 |
Structure Of The Azide-Inhibited Form Of Cytochrome C Peroxidase From Obligate Human Pathogenic Bacterium Neisseria Gonorrhoeae
Organism: Neisseria gonorrhoeae
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-02-19 Classification: ELECTRON TRANSPORT Ligands: HEC, CA, AZI |
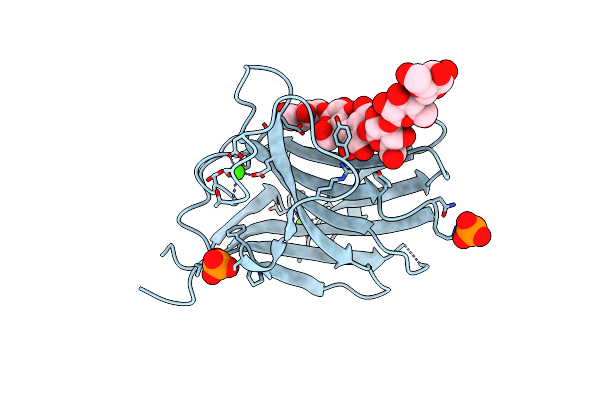 |
Family 11 Carbohydrate-Binding Module From Clostridium Thermocellum In Complex With Beta-1,3-1,4-Mixed-Linked Tetrasaccharide
Organism: Clostridium thermocellum (strain atcc 27405 / dsm 1237 / nbrc 103400 / ncimb 10682 / nrrl b-4536 / vpi 7372)
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2020-02-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, PO4 |
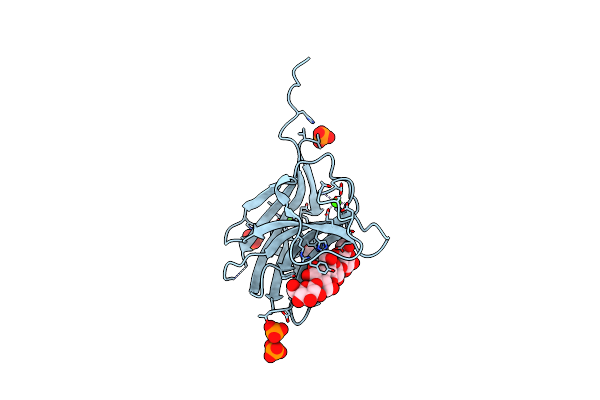 |
Family 11 Carbohydrate-Binding Module From Clostridium Thermocellum In Complex With Beta-1,3-1,4-Mixed-Linked Tetrasaccharide
Organism: Hungateiclostridium thermocellum atcc 27405
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2020-02-05 Classification: SUGAR BINDING PROTEIN Ligands: CA, ACT, PO4, GLC |
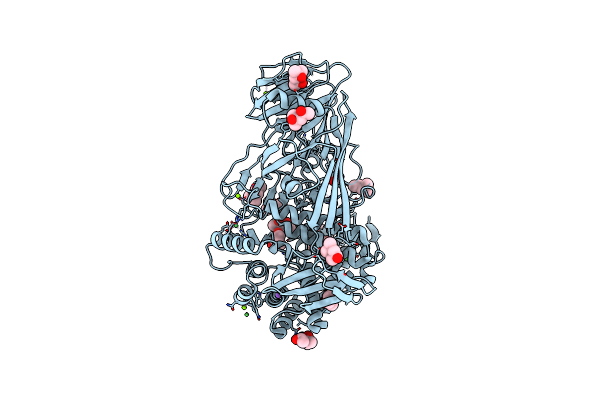 |
Glycoside Hydrolase Family 81 From Clostridium Thermocellum (Ctlam81A), Mutant E515A
Organism: Clostridium thermocellum
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2019-03-13 Classification: HYDROLASE Ligands: CA, MPD, CL, NA, ACT, MG, NI |

