Search Count: 129
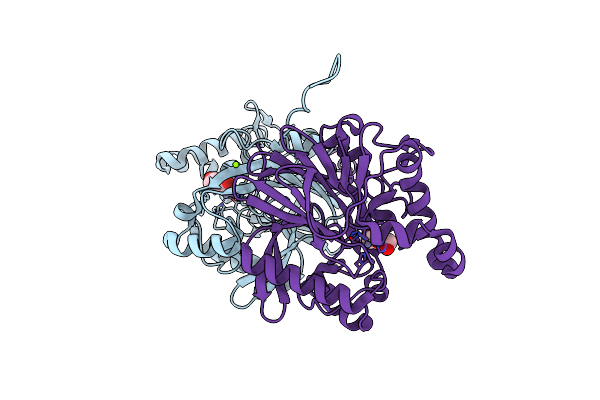 |
The Molecular Basis For The Functional Evolution Of An Organophosphate Hydrolysing Enzyme
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2019-01-16 Classification: HYDROLASE Ligands: ZN, MG, PEG |
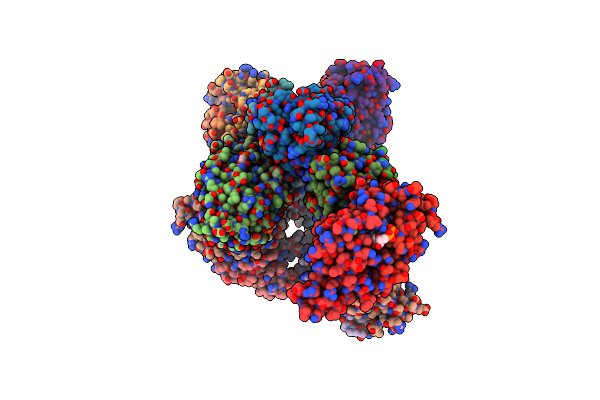 |
Structure Of The Fad Binding Protein Msmeg_5243 From Mycobacterium Smegmatis
Organism: Mycobacterium smegmatis (strain atcc 700084 / mc(2)155)
Method: X-RAY DIFFRACTION Resolution:2.69 Å Release Date: 2018-12-26 Classification: FAD-BINDING PROTEIN Ligands: FAD, CL |
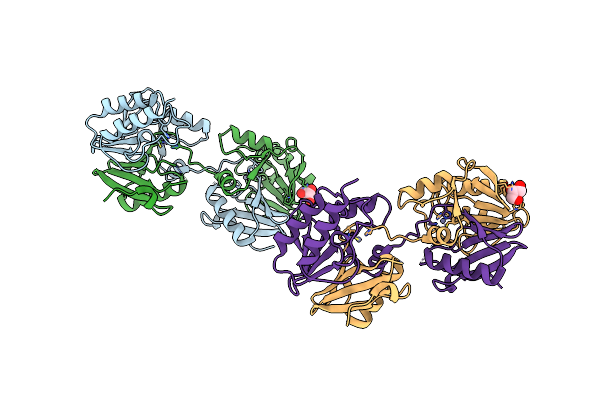 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2018-11-21 Classification: HYDROLASE Ligands: ZN, GOL |
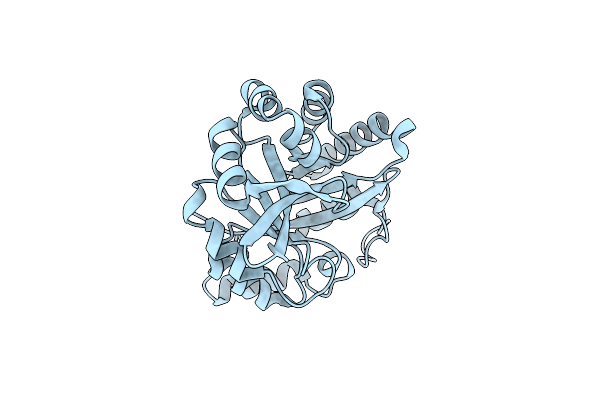 |
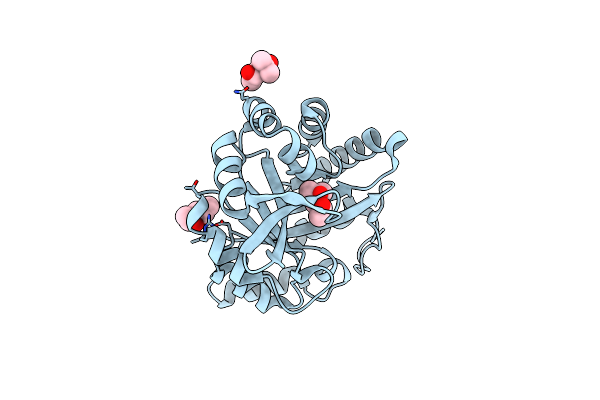
Directed Evolutionary Changes In Kemp Eliminase Ke07 - Crystal 18 Design Trp50Ala Mutant
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.43 Å Release Date: 2018-08-01 Classification: LYASE |
 |
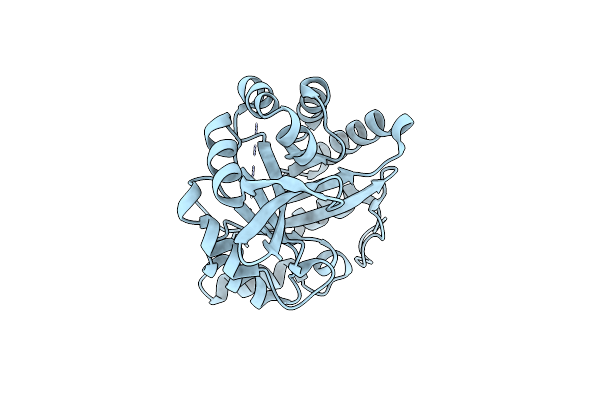
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2018-08-01 Classification: LYASE Ligands: MPD |
 |
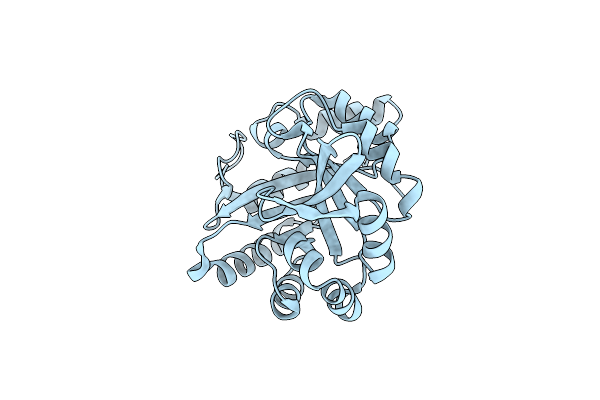
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2018-08-01 Classification: LYASE |
 |
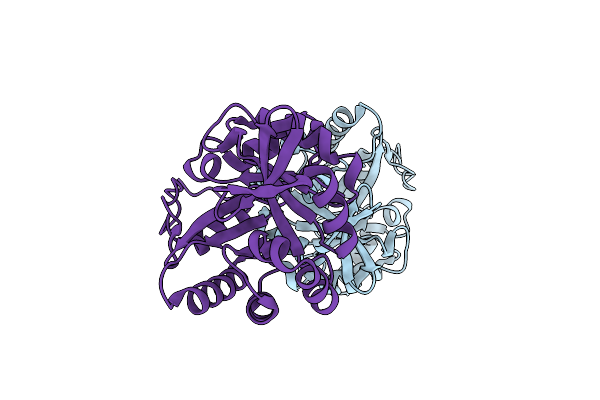
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.64 Å Release Date: 2018-08-01 Classification: LYASE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.61 Å Release Date: 2018-08-01 Classification: LYASE |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2018-08-01 Classification: LYASE Ligands: TRS |
 |
Directed Evolutionary Changes In Kemp Eliminase Ke07 - Crystal 27 Round 7-2
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2018-08-01 Classification: LYASE Ligands: GOL, 6VP, SO4 |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.68 Å Release Date: 2018-08-01 Classification: LYASE Ligands: 6VP, B3P, PEG, TRS |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.89 Å Release Date: 2018-08-01 Classification: LYASE Ligands: PEG, 6VP |
 |
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2018-08-01 Classification: LYASE Ligands: MPD, H5J |
 |
Low-Resolution Structure Of Cyclohexadienyl Dehydratase From Pseudomonas Aeruginosa In Space Group P4322.
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2017-12-13 Classification: LYASE Ligands: ACT |
 |
Organism: Unidentified
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2017-12-06 Classification: Solute Binding Protein |
 |
Crystal Structure Of The Ancestral Amino Acid-Binding Protein Anccdt-1, A Precursor Of Cyclohexadienyl Dehydratase
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.59 Å Release Date: 2017-09-06 Classification: TRANSPORT PROTEIN Ligands: ARG |
 |
Organism: Culex quinquefasciatus
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2017-08-16 Classification: HYDROLASE Ligands: EPE, MLI |
 |
Structure Of F420 Binding Protein, Msmeg_6526, From Mycobacterium Smegmatis With F420 Bound
Organism: Mycobacterium smegmatis
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2017-08-09 Classification: OXIDOREDUCTASE Ligands: F42, MPD, MRD, NA |
 |
Crystal Structure Of The Cyclohexadienyl Dehydratase-Like Solute-Binding Protein Sar11_1068 From Candidatus Pelagibacter Ubique.
Organism: Pelagibacter ubique
Method: X-RAY DIFFRACTION Resolution:1.57 Å Release Date: 2017-08-02 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of Sar11_1068 Bound To A Sulfobetaine (3-(1-Methylpiperidinium-1-Yl)Propane-1-Sulfonate)
Organism: Pelagibacter ubique (strain htcc1062)
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2017-07-05 Classification: LYASE Ligands: KH2, SO4 |

