Search Count: 15
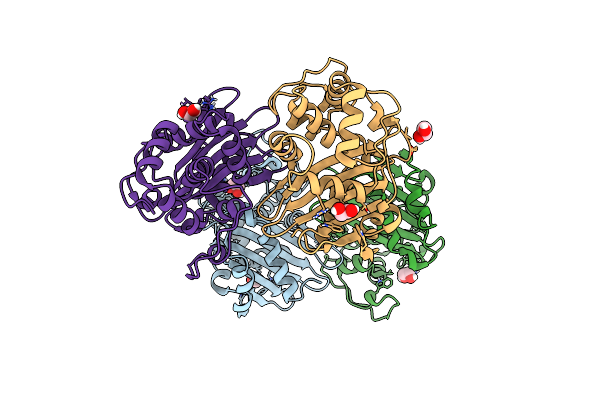 |
Organism: Enterobacter asburiae
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2021-09-08 Classification: HYDROLASE Ligands: EDO, CL |
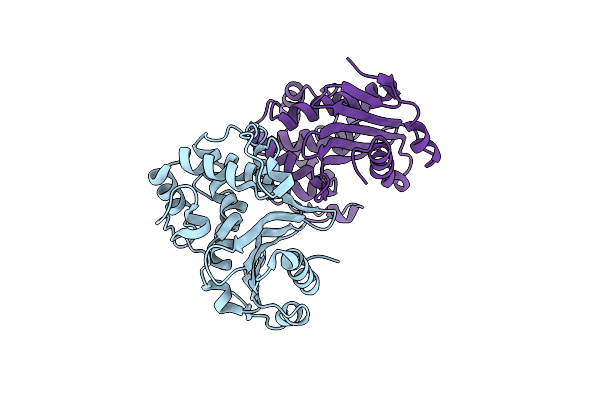 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:3.20 Å Release Date: 2018-08-01 Classification: ANTIBIOTIC |
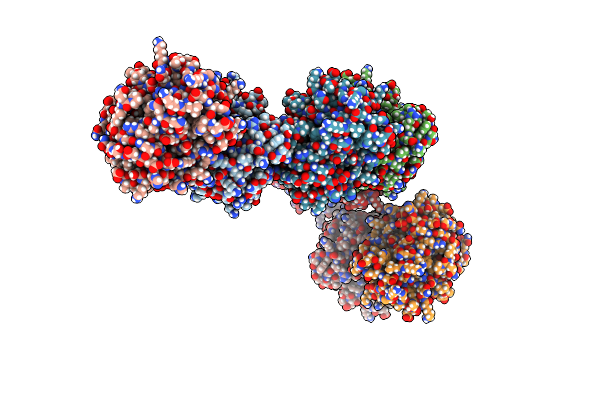 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.55 Å Release Date: 2018-07-18 Classification: HYDROLASE Ligands: CL |
 |
Organism: Enterobacter cloacae
Method: X-RAY DIFFRACTION Resolution:2.07 Å Release Date: 2017-10-18 Classification: ANTIBIOTIC Ligands: CL, MG, PGE |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2017-08-02 Classification: ANTIBIOTIC Ligands: CL, SO4 |
 |
Organism: Klebsiella pneumoniae
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2017-08-02 Classification: ANTIBIOTIC Ligands: CL, NA |
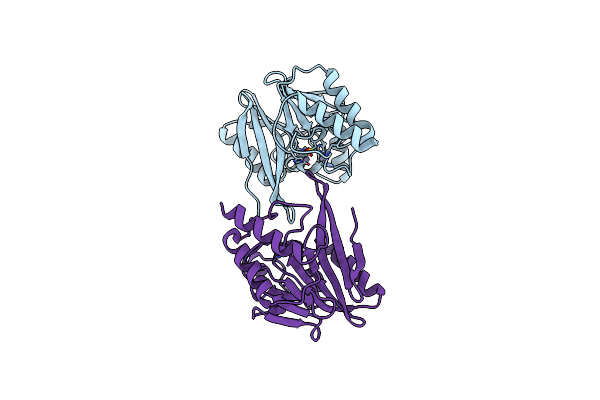 |
W228R-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN, MG |
 |
W228A-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN |
 |
W228Y-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN, CA |
 |
Y233A-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN |
 |
W228S-Investigation Of The Impact From Residues W228 And Y233 In The Metallo-Beta-Lactamase Gim-1
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.81 Å Release Date: 2015-12-23 Classification: HYDROLASE Ligands: ZN |
 |
Vim-2-Nat, Discovery Of Novel Inhibitor Scaffolds Against The Metallo- Beta-Lactamase Vim-2 By Spr Based Fragment Screening
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: OH, ZN, CL |
 |
Vim-2-Ox, Discovery Of Novel Inhibitor Scaffolds Against The Metallo- Beta-Lactamase Vim-2 By Spr Based Fragment Screening
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: OH, ZN, CL |
 |
Vim-2-1, Discovery Of Novel Inhibitor Scaffolds Against The Metallo- Beta-Lactamase Vim-2 By Spr Based Fragment Screening
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: ZN, RHU, CL, OH |
 |
Vim-2-2, Discovery Of Novel Inhibitor Scaffolds Against The Metallo- Beta-Lactamase Vim-2 By Spr Based Fragment Screening
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2015-11-04 Classification: HYDROLASE Ligands: ZN, WL3, OH, CL |

