Search Count: 18
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, CL, NA, DMS, EDO |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar Mg-132.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 150 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 75 Micromolar X77.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: CL, X77, EDO, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer R.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: EDO, CL, X77, DMS, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 500 Micromolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.47 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: DMS, EDO, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Millimolar X77 Enantiomer S.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: 9M5, EDO, ACT, DMS, CL, NA |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm Mg-132, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ALD, EDO, PEG, NA, CL |
 |
Crystal Structure Of The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 Obtained In Presence Of 5 Mm X77, From An "Old" Crystal.
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2024-05-01 Classification: VIRAL PROTEIN Ligands: ACT, EDO, CL, X77, NA |
 |
The 1.68-A X-Ray Crystal Structure Of Sporosarcina Pasteurii Urease Inhibited By Thiram And Bound To Dimethylditiocarbamate
Organism: Sporosarcina pasteurii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2023-11-01 Classification: HYDROLASE Ligands: EDO, SO4, IS9, NI, OH |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: MPD, RB, BA |
 |
Organism: Bacillus cereus atcc 14579
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2023-02-22 Classification: TRANSPORT PROTEIN Ligands: RB, CA, MPD, CL |
 |
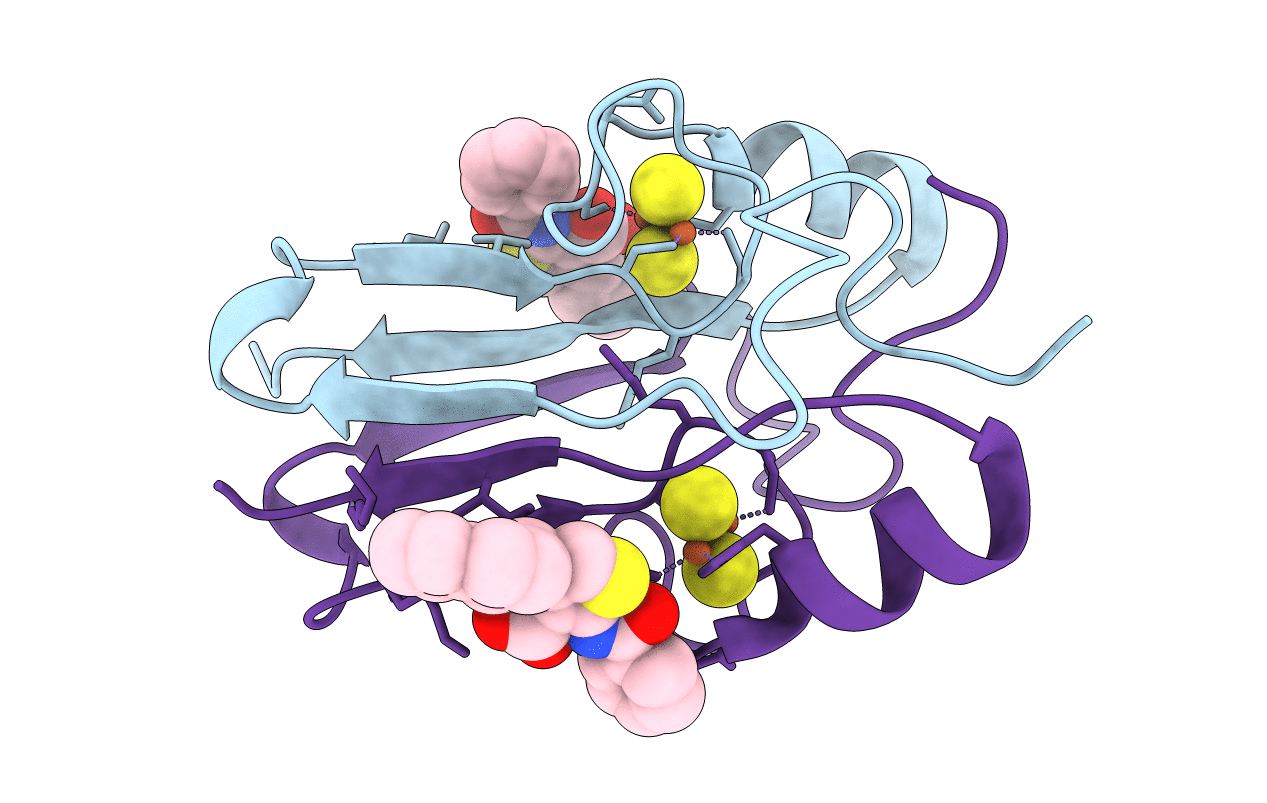
Crystal Structure Of X77 Bound To The Main Protease (3Clpro/Mpro) Of Sars-Cov-2 In Spacegroup P2(1)2(1)2(1).
Organism: Severe acute respiratory syndrome coronavirus 2
Method: X-RAY DIFFRACTION Resolution:1.66 Å Release Date: 2022-09-07 Classification: VIRAL PROTEIN Ligands: X77, EDO, CL, DMS, NA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2022-05-25 Classification: METAL BINDING PROTEIN Ligands: FES, 49I, 4OY |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2022-05-25 Classification: METAL BINDING PROTEIN Ligands: FES, 49I |
 |
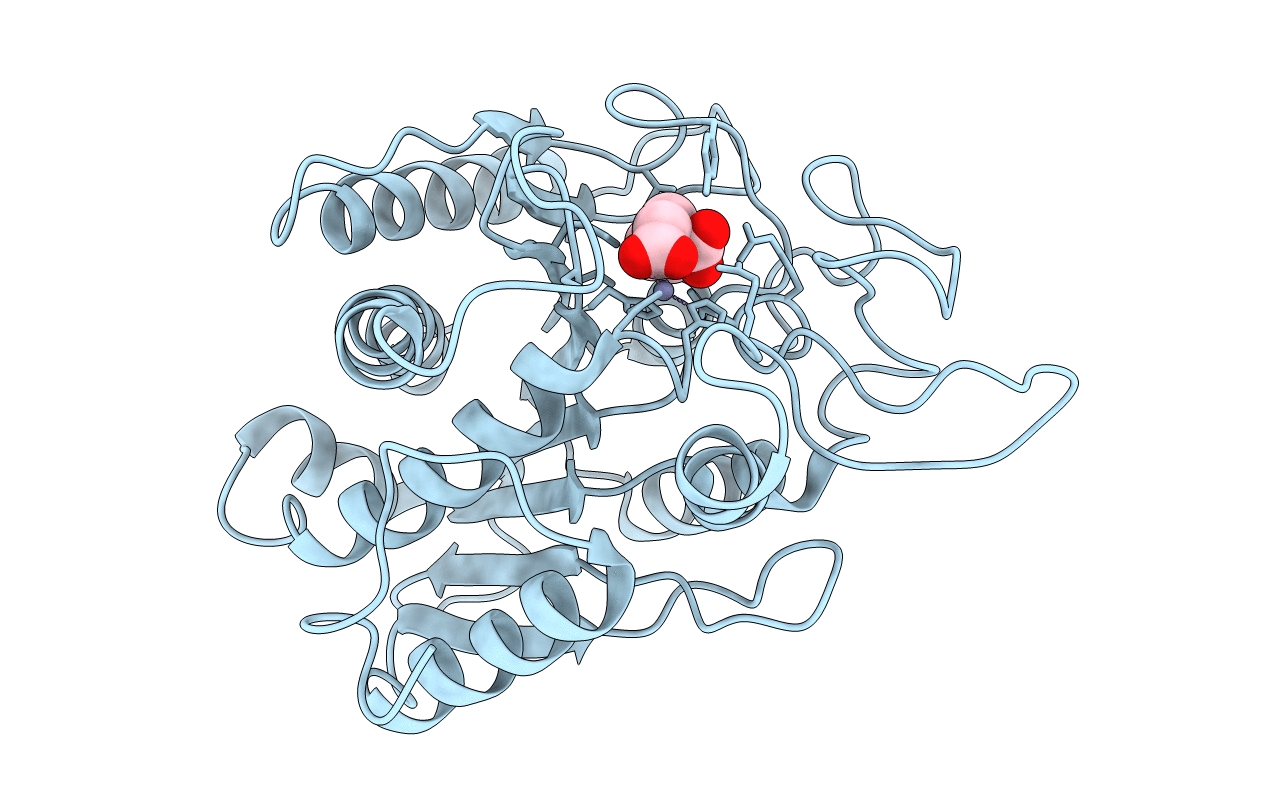
Adaptive Protein Evolution Grants Organismal Fitness By Improving Catalysis And Flexibility
Organism: Bacillus cereus
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2008-12-09 Classification: HYDROLASE Ligands: ZN |
 |
Crystal Structure Of The Complex Between Carboxypeptidase A And The Biproduct Analog Inhibitor L-Benzylsuccinate At 2.0 Angstroms Resolution
Organism: Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 1994-01-31 Classification: HYDROLASE(C-TERMINAL PEPTIDASE) Ligands: ZN, BZS |

