Search Count: 33
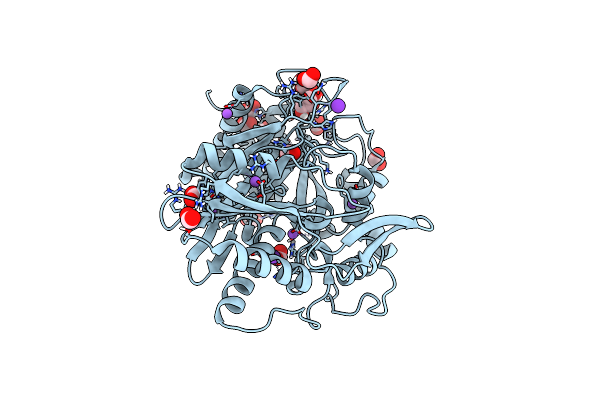 |
Crystal Structure Of The Condensation Domain Tombc From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, GOL, NA, K |
 |
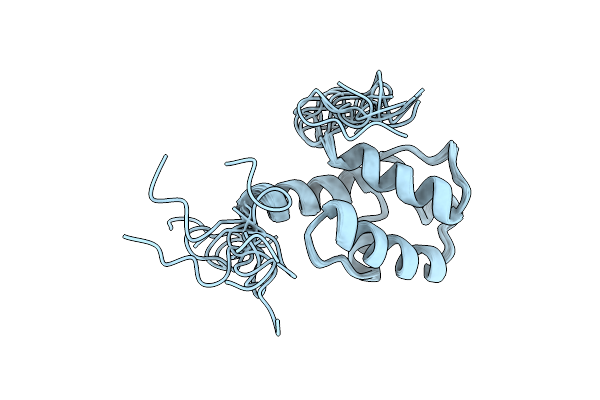
Solution Nmr Structure Of The Peptidyl Carrier Domain Tomapcp From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Solution Nmr Structure Of The Peptidyl Carrier Domain Tomapcp From The Tomaymycin Non-Ribosomal Peptide Synthetase In Its Substrate-Loaded State
Organism: Streptomyces regensis
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Solution Nmr Structure Of The Novel Adaptor Domain Tombn91 From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: SOLUTION NMR Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN |
 |
Semet Derivative Structure Of The Condensation Domain Tombc From The Tomaymycin Non-Ribosomal Peptide Synthetase
Organism: Streptomyces regensis
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2024-06-26 Classification: BIOSYNTHETIC PROTEIN Ligands: FMT, K, GOL |
 |
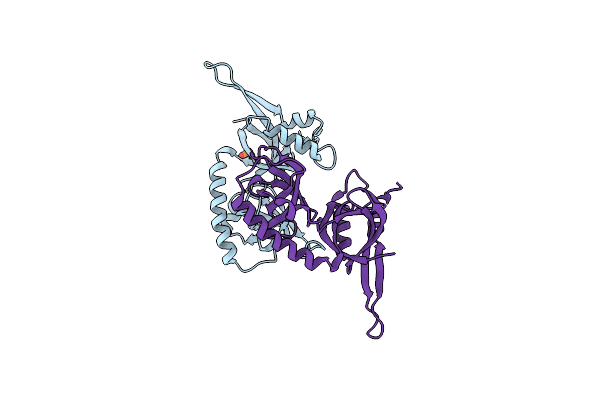
Organism: Severe acute respiratory syndrome coronavirus 2
Method: SOLUTION NMR Release Date: 2022-12-28 Classification: VIRAL PROTEIN |
 |
Organism: Drosophila melanogaster
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-05-04 Classification: PROTEIN BINDING Ligands: PO4 |
 |
Organism: Pyrococcus furiosus (strain atcc 43587 / dsm 3638 / jcm 8422 / vc1), Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2021-10-27 Classification: RIBOSOMAL PROTEIN Ligands: ACT, NA, CA, CL |
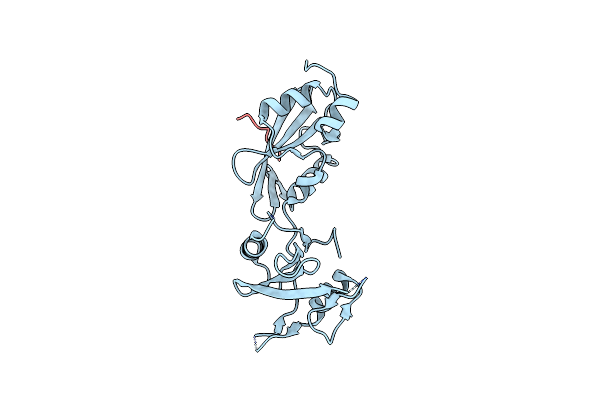 |
Structure Of The Phospholipase C Gamma 1 Tsh2 Domain In Complex With A Phosphorylated Kshv Pk15 Peptide
Organism: Homo sapiens, Human herpesvirus 8 strain gk18
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2021-06-09 Classification: VIRAL PROTEIN |
 |
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2021-02-03 Classification: RIBOSOMAL PROTEIN |
 |
Structure Of The N-Sh2 Domain Of The Human Tyrosine-Protein Phosphatase Non-Receptor Type 11 In Complex With The Phosphorylated Immune Receptor Tyrosine-Based Inhibitory Motif
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-02-19 Classification: CELL CYCLE |
 |
Structure Of The N-Sh2 Domain Of The Human Tyrosine-Protein Phosphatase Non-Receptor Type 11 In Complex With The Phosphorylated Immune Receptor Tyrosine-Based Switch Motif
Organism: Homo sapiens, Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.89 Å Release Date: 2020-02-19 Classification: CELL CYCLE |
 |
 |
Organism: Homo sapiens
Method: SOLUTION NMR Release Date: 2020-02-05 Classification: PEPTIDE BINDING PROTEIN |
 |
Structure Of Asf1-H3:H4-Rtt109-Vps75 Histone Chaperone-Lysine Acetyltransferase Complex With The Histone Substrate.
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Xenopus laevis
Method: SOLUTION NMR, SOLUTION SCATTERING Release Date: 2019-07-31 Classification: CHAPERONE |
 |
Organism: Saccharomyces cerevisiae (strain atcc 204508 / s288c), Saccharomyces cerevisiae s288c
Method: SOLUTION NMR Release Date: 2017-12-27 Classification: TRANSFERASE |
 |
 |
Solution Structure Of A Double Base-Pair Inversion Mutant Of Murine Tumour Necrosis Factor Alpha Cde-23 Rna
|
 |
 |

