Search Count: 164
 |
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN |
 |
Human Delta Opioid Receptor Complex With Mini-Gi And Agonist Dadle And Allosteric Modulator Mips3614
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CVG |
 |
Human Delta Opioid Receptor Complex With Mini-Gi And Agonist Dadle And Allosteric Modulator Mips3983
Organism: Homo sapiens, Lama glama
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: MEMBRANE PROTEIN Ligands: A1CU8 |
 |
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Release Date: 2024-12-11 Classification: OXIDOREDUCTASE Ligands: A1BH7 |
 |
Organism: Rattus norvegicus, Gallus gallus, Bos taurus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2021-07-07 Classification: CELL CYCLE Ligands: GTP, MG, CA, GDP, MES, GOL, RYK, IMD, ACP |
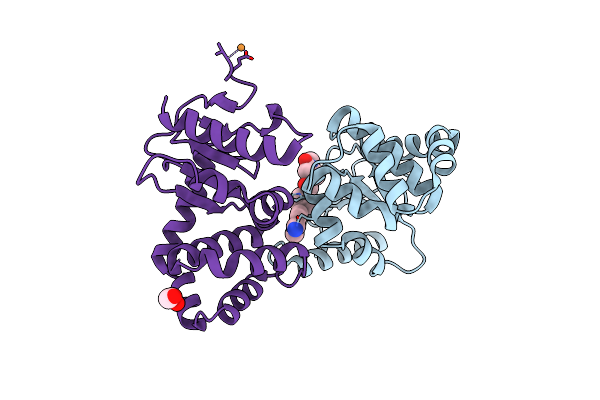 |
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product F1 (Methylpiperazinone 6)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.01 Å Release Date: 2020-07-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O7P, CU, EDO |
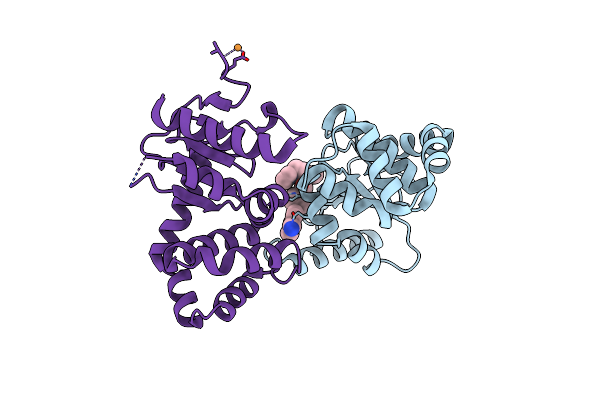 |
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product G6 (Pyrazole 9)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-07-01 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OAV, CU |
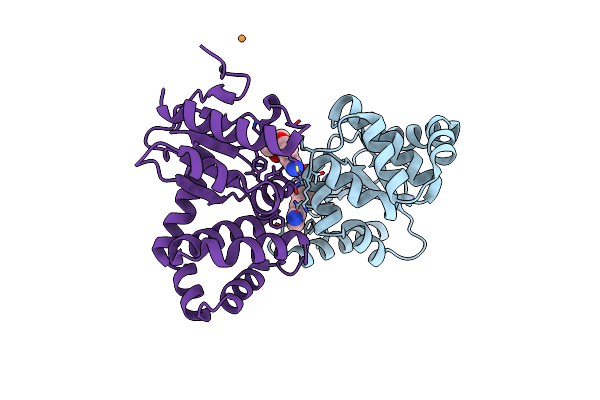 |
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2020-06-24 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: KFS, CU |
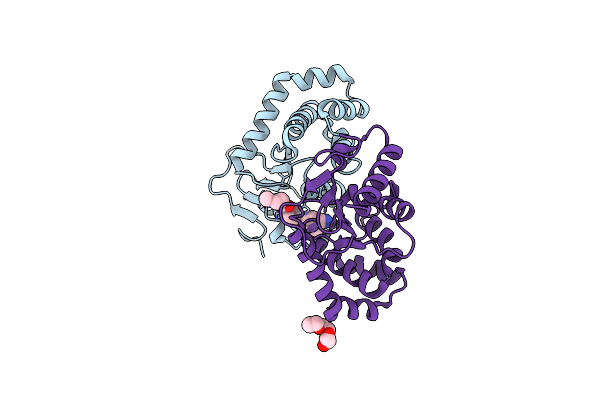 |
Crystal Structure Of Ecdsba In A Complex With Purified Methylpiperazinone 6
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2020-05-20 Classification: OXIDOREDUCTASE/OXIDOREDUCTASE INHIBITOR Ligands: O7P, PGE |
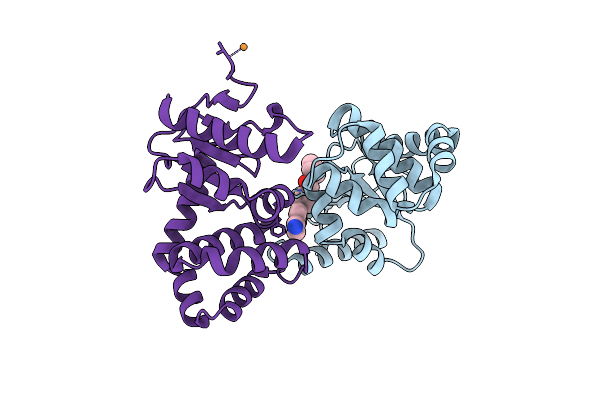 |
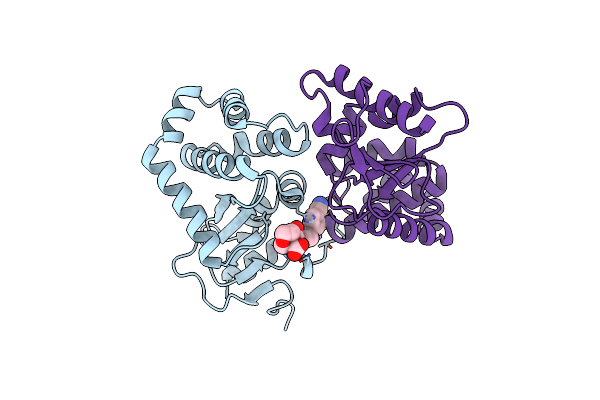
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O6Y, CU |
 |
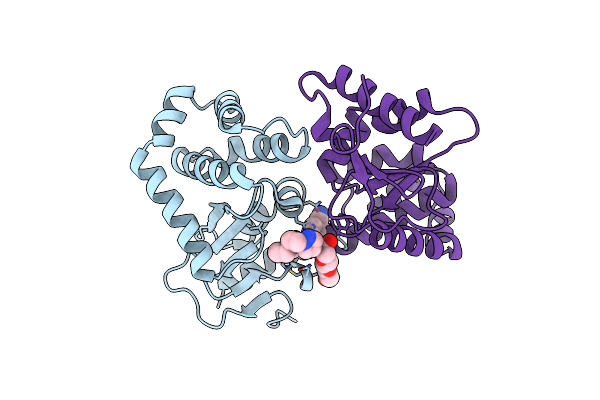
Crystal Structure Of Ecdsba In A Complex With Purified Morpholine Carboxylic Acid 7
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OAJ |
 |
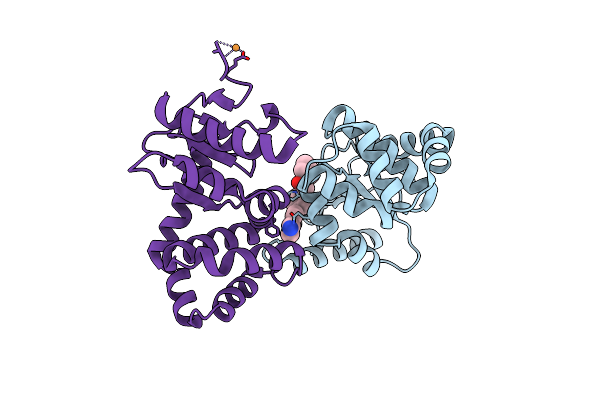
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.96 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OAV, PGE |
 |
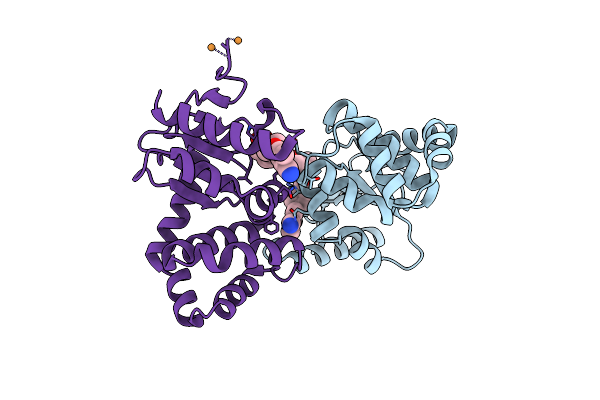
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product H5 (Morpholine 8)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.91 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: O6Y, CU |
 |
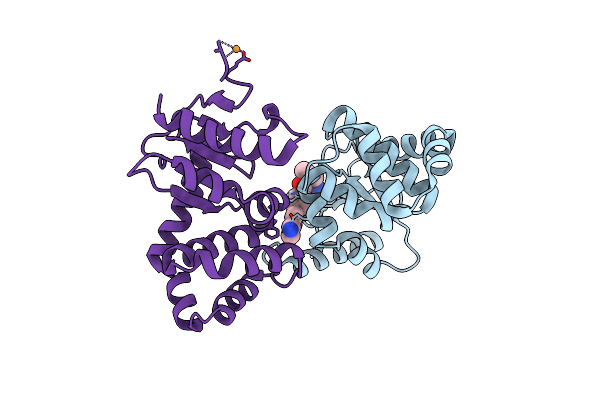
Crystal Structure Of Ecdsba In A Complex With Unpurified Reaction Product A5 (Morpholine Carboxylic Acid 7)
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: OMJ, CU, OAJ |
 |
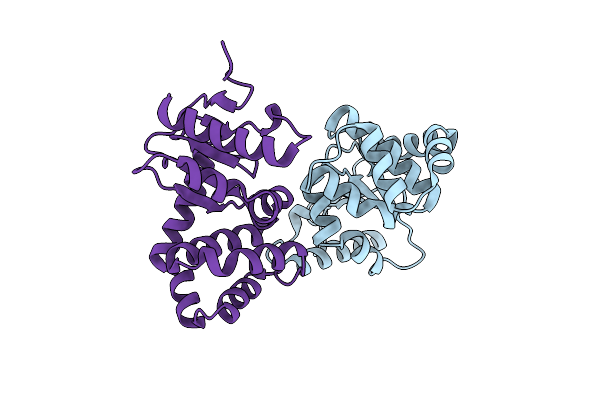
Organism: Escherichia coli (strain k12)
Method: X-RAY DIFFRACTION Resolution:1.93 Å Release Date: 2020-05-06 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: ONY, CU |
 |
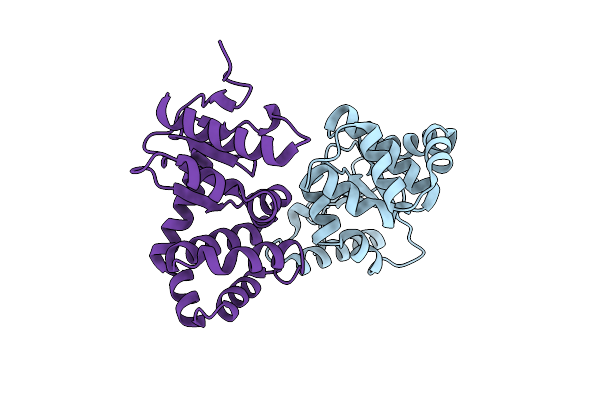
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A1_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.15 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
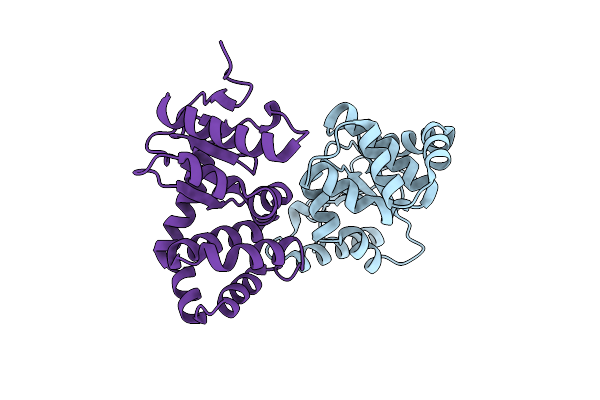
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A10_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.11 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
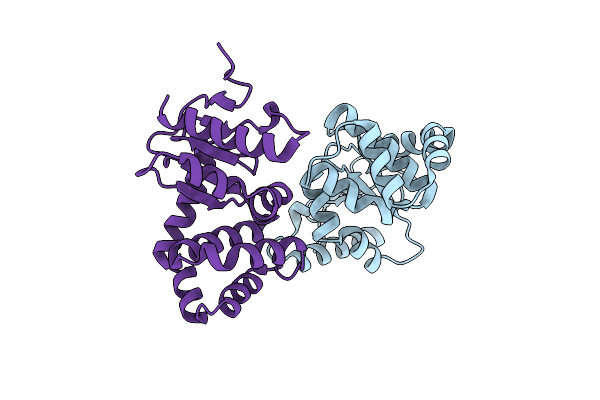
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A11_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.12 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
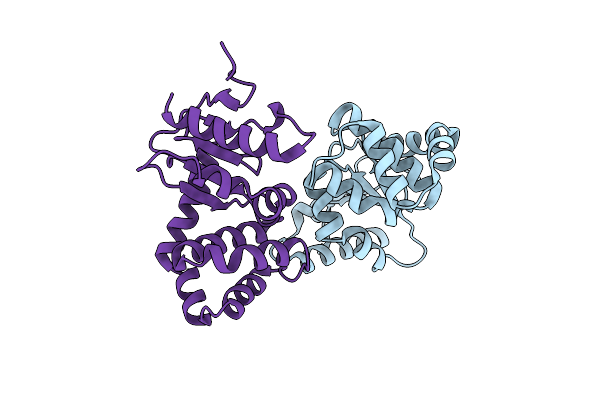
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A12_1)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:2.14 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |
 |
Group Deposition Of Library Data - Crystal Structure Of Ecdsba After Initial Refinement With No Ligand Modelled (Structure A2_3)
Organism: Escherichia coli k-12
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2020-02-05 Classification: OXIDOREDUCTASE |

