Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
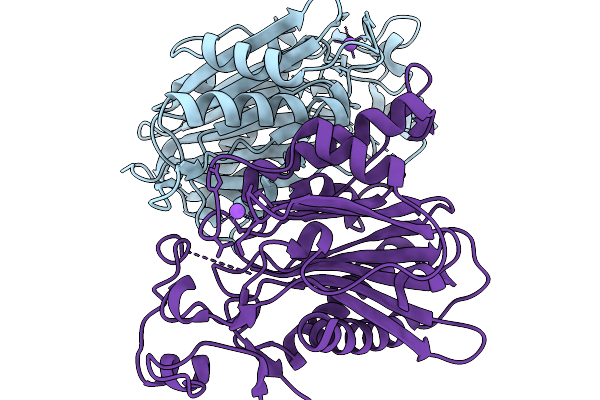 |
Crystal Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 In Its Apo Form At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Release Date: 2025-11-19 Classification: HYDROLASE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PGE |
 |
X-Ray Crystallographic Structure Of The Poly(Hexamethylene Adipamide) (Nylon66) Hydrolase Nyl50 At Room Temperature Bound To Tetraethylene Glycol
Organism: Alphaproteobacteria bacterium
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2025-01-29 Classification: HYDROLASE Ligands: EDO, NA, PG4 |
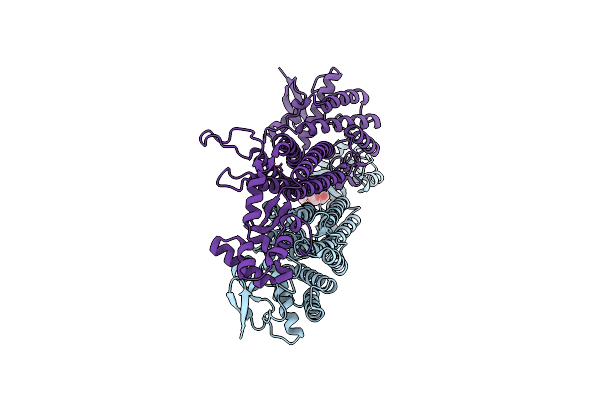 |
Crystal Structure Of Mutant Aspartase From Bacillus Sp. Ym55-1 In The Closed Loop Conformation
Organism: Bacillus sp. ym55-1
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2025-01-15 Classification: LYASE Ligands: PGE, NA |
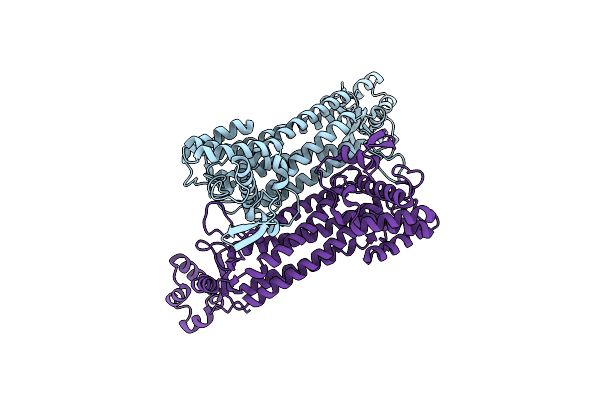 |
Crystal Structure Of Mutant Aspartase From Caenibacillus Caldisaponilyticus In The Closed Loop Conformation
Organism: Caenibacillus caldisaponilyticus
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2025-01-15 Classification: LYASE |
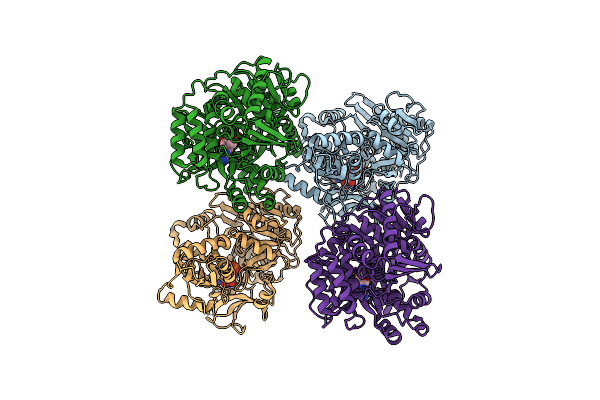 |
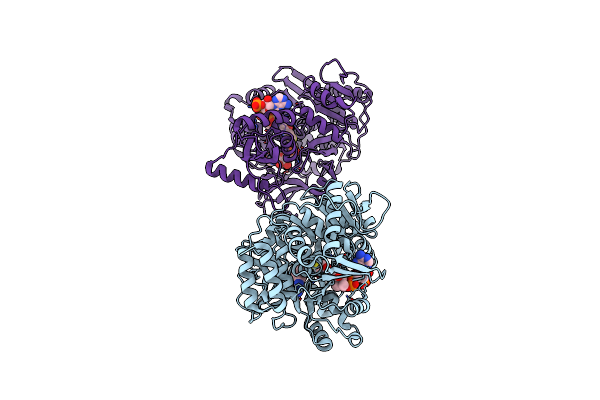
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: AMP, 6R9 |
 |
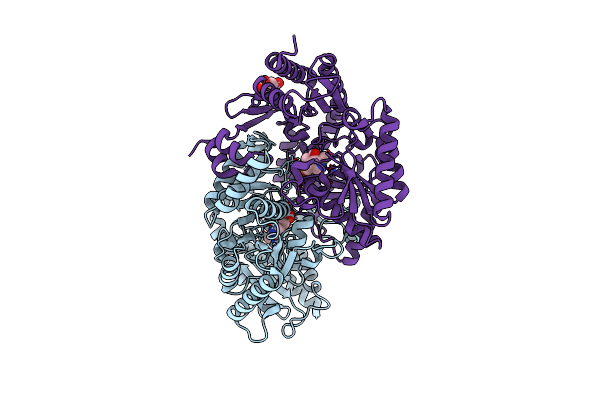
Crystal Structure Of Acyl-Coa Synthetase From Metallosphaera Sedula In Complex With Coenzyme A And Acetyl-Amp
Organism: Metallosphaera sedula dsm 5348
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2023-11-15 Classification: LIGASE Ligands: 6R9, COA |
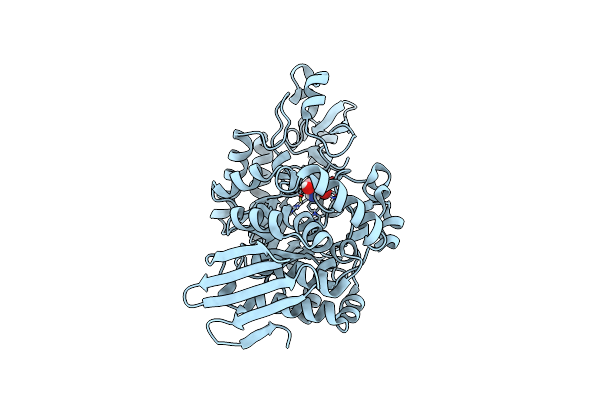 |
Organism: Faecalibaculum rodentium
Method: X-RAY DIFFRACTION Resolution:1.36 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: TRS |
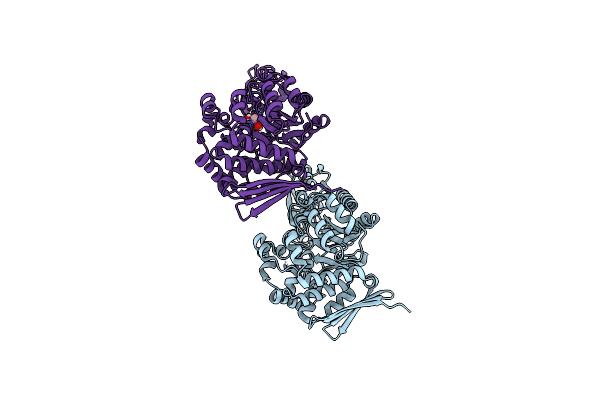 |
Organism: Jeotgalibaca ciconiae
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2023-01-18 Classification: TRANSFERASE Ligands: TRS |
 |
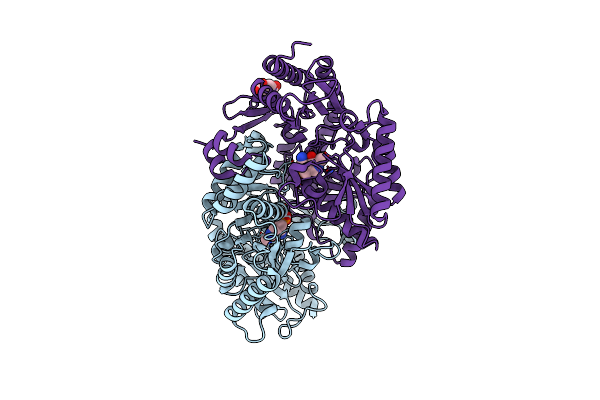
Thermostable Omega Transaminase Pjta-R6 Variant W58G Engineered For Asymmetric Synthesis Of Enantiopure Bulky Amines
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: PLP, SIN |
 |
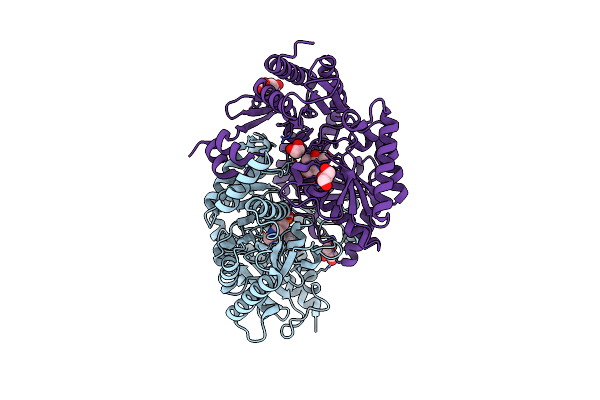
Thermostable Omega Transaminase Pjta-R6 Variant W58M/F86L/R417L Engineered For Asymmetric Synthesis Of Enantiopure Bulky Amines
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-01 Classification: TRANSFERASE Ligands: PMP, SIN |
 |
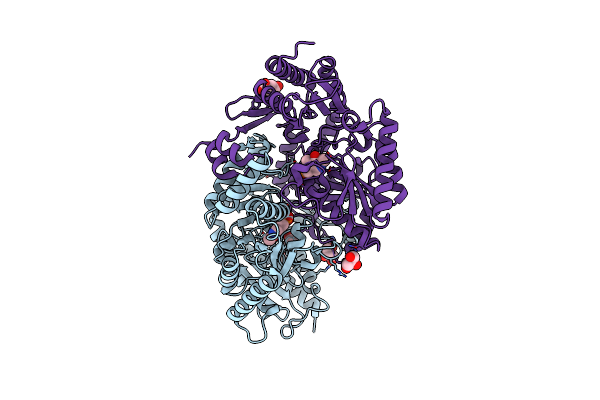
Crystal Structure Of Thermostable Omega Transaminase 4-Fold Mutant From Pseudomonas Jessenii
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: PLP, GOL, SIN |
 |
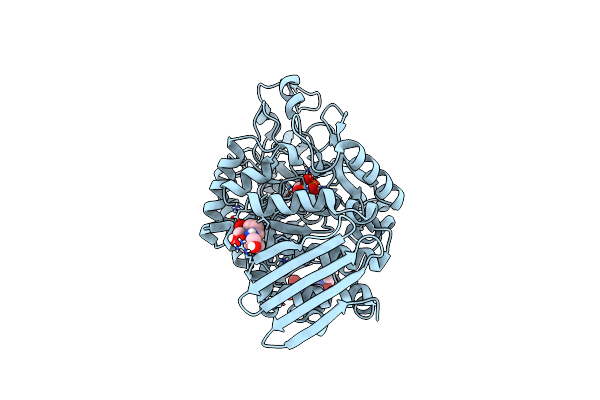
Crystal Structure Of Thermostable Omega Transaminase 6-Fold Mutant From Pseudomonas Jessenii
Organism: Pseudomonas sp.
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2020-07-15 Classification: TRANSFERASE Ligands: PLP, GOL, SIN, NA |
 |
Crystal Structure Of Sucrose 6F-Phosphate Phosphorylase From Ilumatobacter Coccineus
Organism: Ilumatobacter coccineus ym16-304
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2019-08-28 Classification: TRANSFERASE Ligands: PO4, B3P, TRS |
 |
Crystal Structure Of Sucrose 6F-Phosphate Phosphorylase From Thermoanaerobacter Thermosaccharolyticum
Organism: Thermoanaerobacterium thermosaccharolyticum dsm 571
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2019-08-28 Classification: TRANSFERASE Ligands: BTB, SO4, P6G, GOL |