Search Count: 12
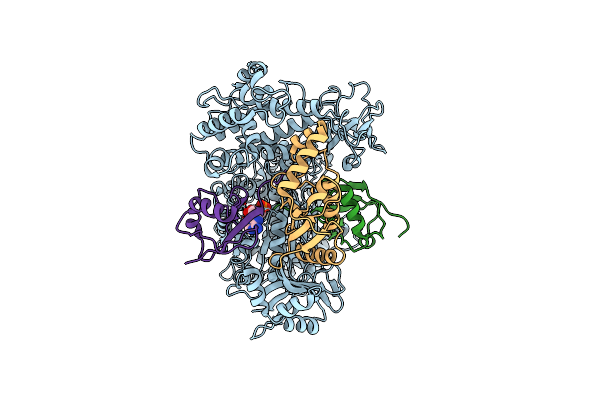 |
Cryo-Em Structure Of A Double Loaded Human Uba7-Ube2L6-Isg15 Thioester Mimetic Complex (Form 2)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: LIGASE Ligands: AMP |
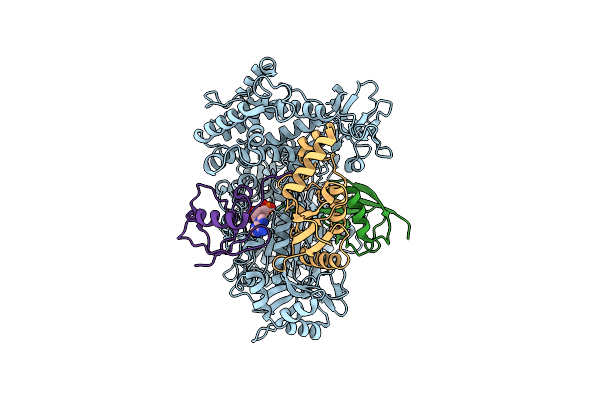 |
Cryo-Em Structure Of A Double Loaded Human Uba7-Ube2L6-Isg15 Thioester Mimetic Complex (Form 1)
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: LIGASE Ligands: AMP |
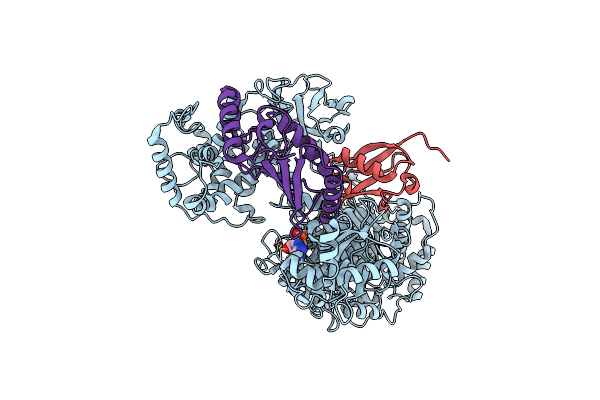 |
Cryo-Em Structure Of A Single Loaded Human Uba7-Ube2L6-Isg15 Adenylate Complex
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: LIGASE Ligands: AMP |
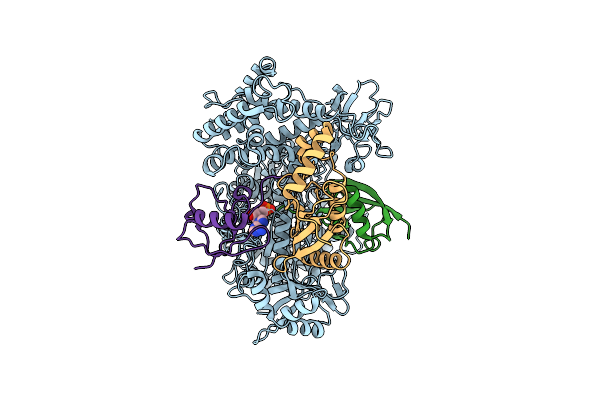 |
Cryo-Em Structure Of A Double Loaded Human Uba7-Ube2L6-Isg15 Thioester Mimetic Complex From A Composite Map
Organism: Homo sapiens
Method: ELECTRON MICROSCOPY Release Date: 2023-10-11 Classification: SIGNALING PROTEIN Ligands: AMP |
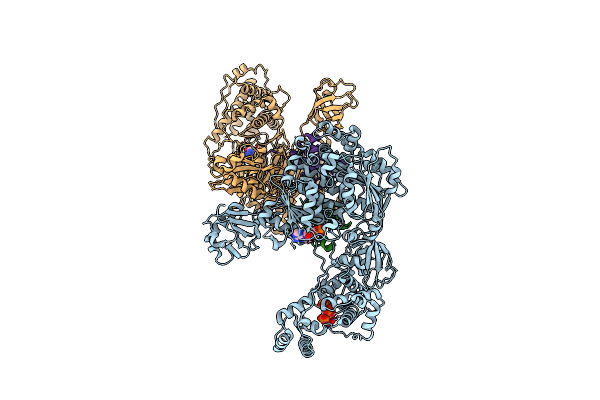 |
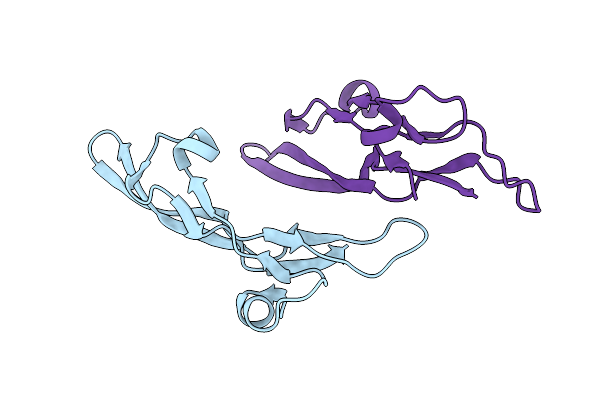
Crystal Structures Of The Bispecific Ubiquitin/Fat10 Activating Enzyme, Uba6
Organism: Homo sapiens, Triticum aestivum
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2022-11-02 Classification: SIGNALING PROTEIN/Ligase Ligands: IHP, AMP |
 |
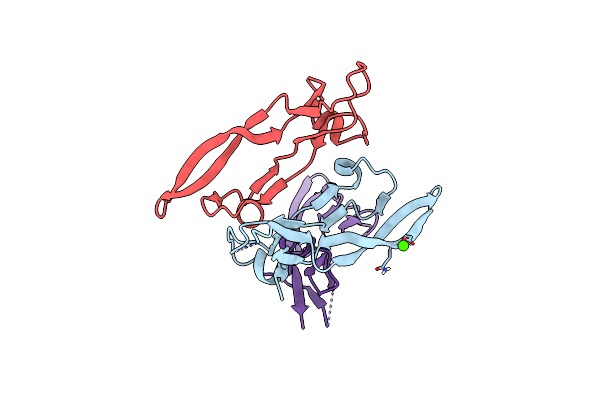
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.82 Å Release Date: 2017-03-01 Classification: CYTOKINE |
 |
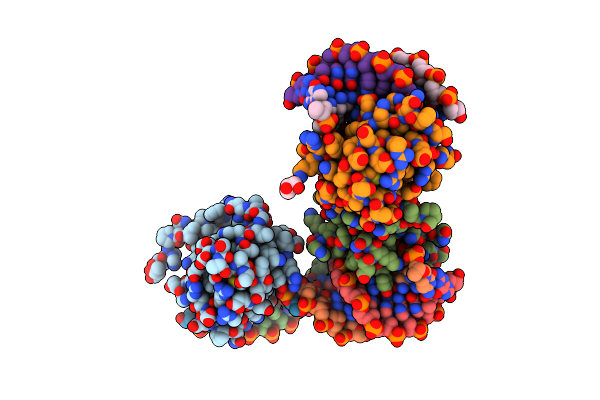
Derivative Of Mouse Tgf-Beta2, With A Deletion Of Residues 52-71 And K25R, R26K, L51R, A74K, C77S, L89V, I92V, K94R T95K, I98V Single Amino Acid Substitutions, Bound To Human Tgf-Beta Type Ii Receptor Ectodomain Residues 15-130
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.88 Å Release Date: 2017-03-01 Classification: TRANSFERASE/CYTOKINE |
 |
Structure Of Tgf-Beta2 Derivative With Deletion Of Residues 52-71 And 10 Single Amino Acid Mutations (Mmtgf-Beta2-7M)
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.75 Å Release Date: 2017-03-01 Classification: CYTOKINE Ligands: CA |
 |
Crystal Structure Of The Dna Binding Domain Of Transcription Factor Fli1 In Complex With An 11-Mer Dna Gaccggaagtg
Organism: Homo sapiens, Endothia gyrosa
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-09-14 Classification: transcription/dna Ligands: PO4, GOL |
 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-Trp In Complex With A 10-Mer Dna Agaggcctct.
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2016-09-14 Classification: dna/antibiotic Ligands: ZN, NA, 6O6 |
 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-Phe In Complex With A 10-Mer Dna Agggtaccct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:2.40 Å Release Date: 2016-09-14 Classification: dna/antibiotic Ligands: 6O7, ZN |
 |
Crystal Structure Of Mithramycin Analogue Mtm Sa-Phe In Complex With A 10-Mer Dna Agggatccct
Organism: Synthetic construct
Method: X-RAY DIFFRACTION Resolution:3.10 Å Release Date: 2016-09-14 Classification: DNA/antibiotic Ligands: 6O7, ZN |

