Search Count: 11
 |
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2017-05-24 Classification: LIGASE Ligands: 8Z7 |
 |
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.37 Å Release Date: 2017-05-24 Classification: HYDROLASE Ligands: 8ZA |
 |
Organism: Enterobacteria phage t4, Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2017-05-24 Classification: Ligase / protein binding Ligands: 8ZD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2017-05-24 Classification: Ligase / protein binding |
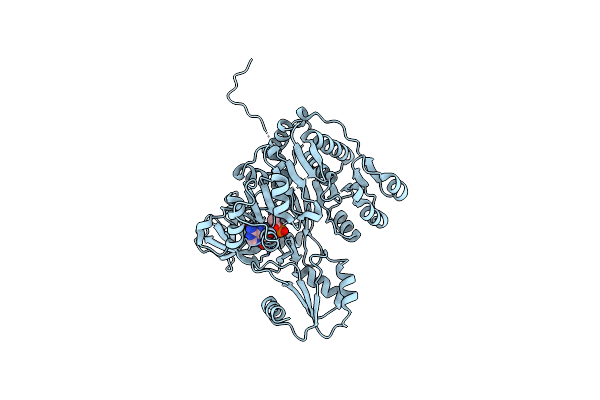 |
Crystal Structure Of Vibrio Cholerae Adenylation Domain Alme In Complex With Glycyl-Adenosine-5'-Phosphate
Organism: Vibrio cholerae serotype o1
Method: X-RAY DIFFRACTION Resolution:2.26 Å Release Date: 2014-12-31 Classification: LIGASE Ligands: GAP |
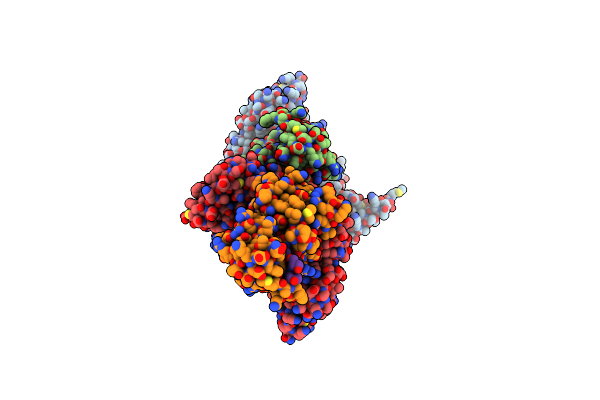 |
Novel Modifications On C-Terminal Domain Of Rna Polymerase Ii Can Fine- Tune The Phosphatase Activity Of Ssu72.
Organism: Drosophila melanogaster, Synthetic
Method: X-RAY DIFFRACTION Resolution:2.35 Å Release Date: 2013-08-07 Classification: HYDROLASE Ligands: PO4 |
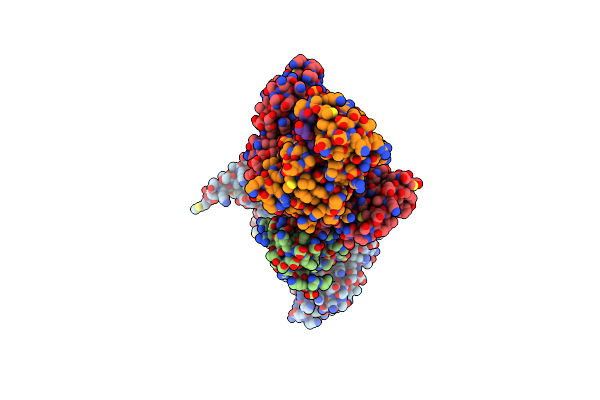 |
Novel Modifications On C-Terminal Domain Of Rna Polymerase Ii Can Fine-Tune The Phosphatase Activity Of Ssu72
Organism: Drosophila melanogaster, Synthetic
Method: X-RAY DIFFRACTION Resolution:2.58 Å Release Date: 2013-08-07 Classification: HYDROLASE Ligands: PO4 |
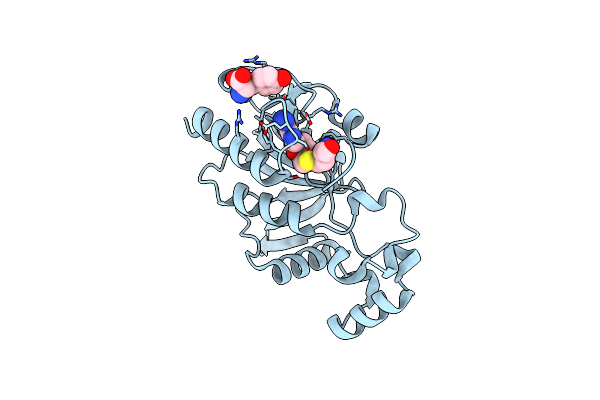 |
Crystal Structure Of Archaeoglobus Fulgidus Nep1 Bound To S-Adenosylhomocysteine
Organism: Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.45 Å Release Date: 2011-01-19 Classification: TRANSFERASE Ligands: TYR, SAH |
 |
Crystal Structure Of Saccharomyces Cerevisiae Nep1/Emg1 Bound To S-Adenosylhomocysteine
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2010-12-01 Classification: RIBOSOMAL PROTEIN Ligands: SAH, GOL |
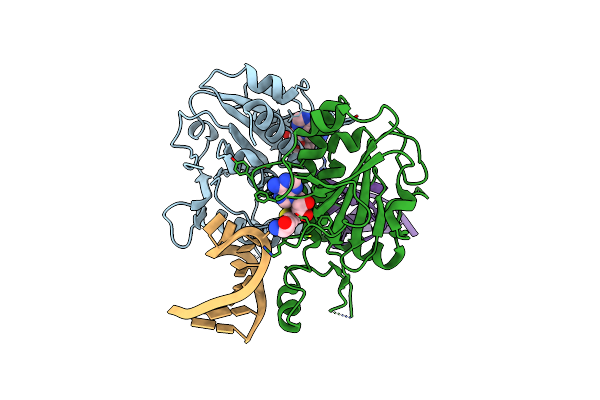 |
Crystal Structure Of Saccharomyces Cerevisiae Nep1/Emg1 Bound To S-Adenosylhomocysteine And 2 Molecules Of Cognate Rna
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:3.00 Å Release Date: 2010-12-01 Classification: RIBOSOMAL PROTEIN Ligands: SAH, MG |
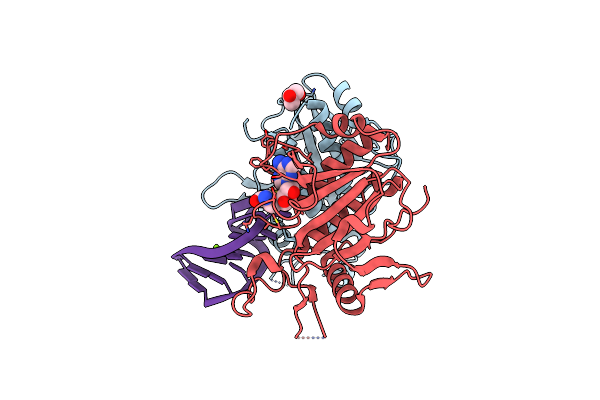 |
Crystal Structure Of Saccharomyces Cerevisiae Nep1/Emg1 Bound To S-Adenosylhomocysteine And 1 Molecule Of Cognate Rna
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2010-12-01 Classification: RIBOSOMAL PROTEIN Ligands: GOL, SAH, MG, CL |

