Search Count: 25
 |
Domain 1 Of Starch Adherence System Protein 20 (Sas20) From Ruminococcus Bromii
Organism: Ruminococcus bromii l2-63
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2022-04-13 Classification: SUGAR BINDING PROTEIN Ligands: PEG, PGE |
 |
Domain 1 Of Starch Adherence System Protein 20 (Sas20) From Ruminococcus Bromii With Maltotriose
Organism: Ruminococcus bromii l2-63
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2022-04-13 Classification: SUGAR BINDING PROTEIN Ligands: EDO, CL, ZN |
 |
Organism: Ruminococcus bromii l2-63
Method: X-RAY DIFFRACTION Resolution:1.67 Å Release Date: 2022-04-13 Classification: SUGAR BINDING PROTEIN Ligands: ACT, GOL |
 |
Organism: Bacteroides intestinalis
Method: X-RAY DIFFRACTION Resolution:1.74 Å Release Date: 2019-12-25 Classification: HYDROLASE Ligands: PEG, EDO, SO4, CL |
 |
Organism: Bacteroides intestinalis dsm 17393
Method: X-RAY DIFFRACTION Resolution:1.71 Å Release Date: 2019-10-16 Classification: HYDROLASE Ligands: EDO |
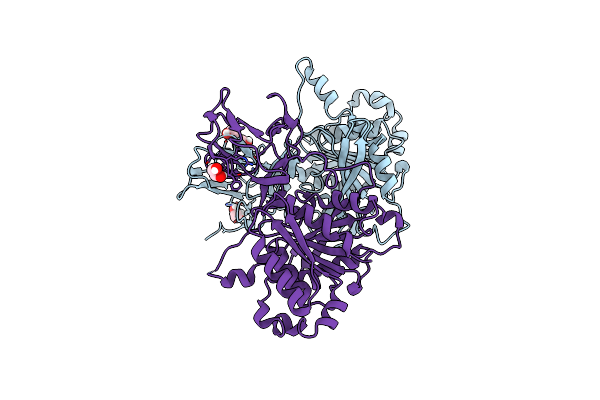 |
Organism: Bacteroides intestinalis dsm 17393
Method: X-RAY DIFFRACTION Resolution:2.24 Å Release Date: 2019-10-16 Classification: HYDROLASE Ligands: EDO |
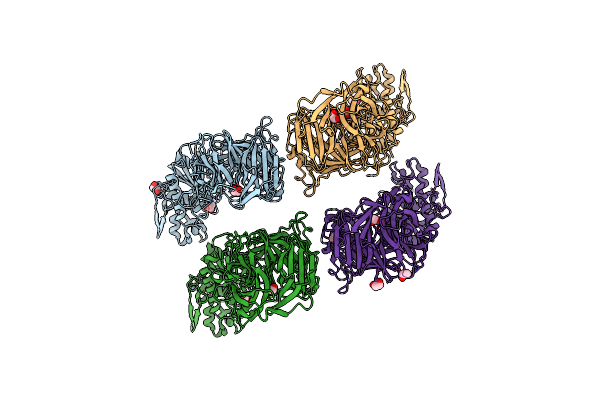 |
Organism: Bacteroides eggerthii
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2019-10-02 Classification: HYDROLASE Ligands: ACT, EDO, NA |
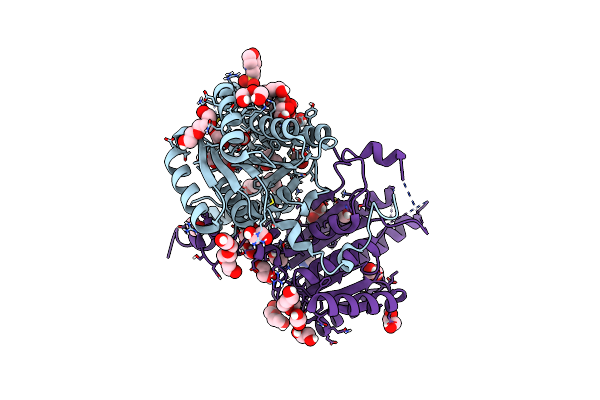 |
Structure Of The Apo-Form Of 20Beta-Hydroxysteroid Dehydrogenase From Bifidobacterium Adolescentis Strain L2-32
Organism: Bifidobacterium adolescentis l2-32
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: IPA, MES, EDO, PGE, PEG, PG4, CL, IMD |
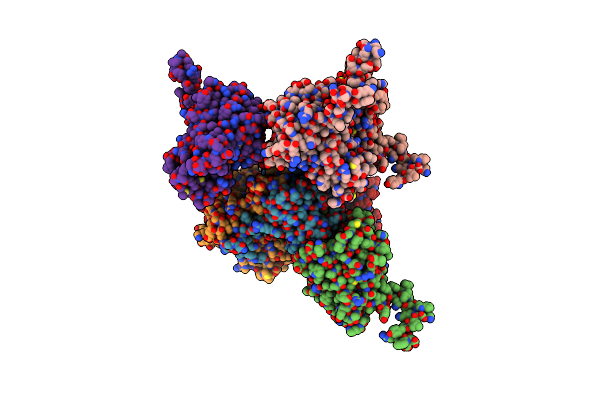 |
Structure Of The Nadh-Bound Form Of 20Beta-Hydroxysteroid Dehydrogenase From Bifidobacterium Adolescentis Strain L2-32
Organism: Bifidobacterium adolescentis l2-32
Method: X-RAY DIFFRACTION Resolution:1.99 Å Release Date: 2019-06-26 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Bacteroides intestinalis dsm 17393
Method: X-RAY DIFFRACTION Resolution:1.98 Å Release Date: 2017-07-19 Classification: HYDROLASE Ligands: CA, IPA |
 |
Organism: Caldanaerobius polysaccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.40 Å Release Date: 2014-10-29 Classification: SUGAR BINDING PROTEIN |
 |
Organism: Caldanaerobius polysaccharolyticus
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2014-10-29 Classification: SUGAR BINDING PROTEIN Ligands: ZN |
 |
Organism: Prevotella bryantii
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2014-08-20 Classification: HYDROLASE Ligands: CA |
 |
Organism: Bacteroides intestinalis
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2014-08-20 Classification: HYDROLASE |
 |
Organism: Bacteroides intestinalis dsm 17393
Method: X-RAY DIFFRACTION Resolution:1.14 Å Release Date: 2014-08-13 Classification: HYDROLASE |
 |
Organism: Caldanaerobius polysaccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2013-02-20 Classification: HYDROLASE Ligands: TRS |
 |
Biochemical And Structural Insights Into Xylan Utilization By The Thermophilic Bacteriumcaldanaerobius Polysaccharolyticus
Organism: Caldanaerobius
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2012-08-22 Classification: TRANSPORT PROTEIN |
 |
Crystal Structure Of The Q121E Mutants Of C.Polysaccharolyticus Cbm16-1 Bound To Cellopentaose
Organism: Caldanaerobius polysaccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.35 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: CA |
 |
Crystal Structure Of The Q121E Mutant Of C.Polysaccharolyticus Cbm16-1 Bound To Mannopentaose
Organism: Caldanaerobius polysaccharolyticus
Method: X-RAY DIFFRACTION Resolution:1.55 Å Release Date: 2010-08-25 Classification: HYDROLASE Ligands: CA, SO4 |
 |
Crystal Structure Analysis Of A Full-Length Mcm Homolog From Methanopyrus Kandleri
Organism: Methanopyrus kandleri av19
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2009-03-03 Classification: HYDROLASE |

