Search Count: 6
 |
Crystal Structure Of Sar11_0769 From 'Candidatus Pelagibacter Ubique' Htcc1062 Bound To A Co-Purified Ligand, Beta-Galactopyranose
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2024-07-17 Classification: TRANSPORT PROTEIN Ligands: GAL |
 |
Crystal Structure Of Sar11_0655 Bound To A Co-Purified Ligand, L-Pyroglutamate
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2024-07-17 Classification: TRANSPORT PROTEIN Ligands: PCA, NA |
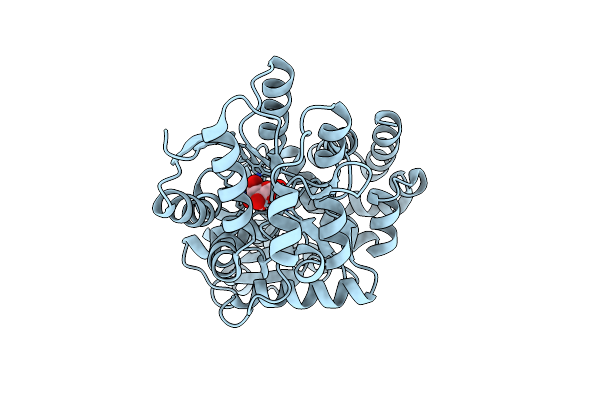 |
Crystal Structure Of The Glucose-Binding Protein Sar11_0769 From "Candidatus Pelagibacter Ubique" Htcc1062 Bound To Glucose
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.86 Å Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: BGC |
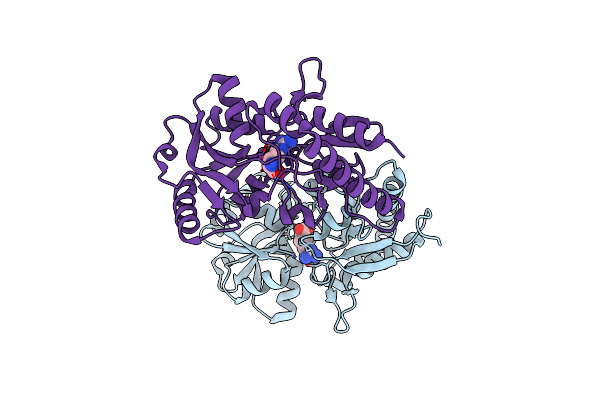 |
Crystal Structure Of The Arginine-/Lysine-Binding Protein Sar11_1210 From 'Candidatus Pelagibacter Ubique' Htcc1062 Bound To Arginine
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.32 Å Release Date: 2023-12-27 Classification: TRANSPORT PROTEIN Ligands: ARG |
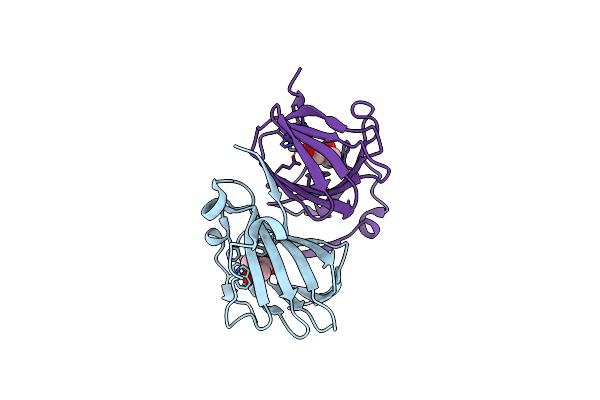 |
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:1.62 Å Release Date: 2023-10-04 Classification: LYASE Ligands: MN, LNI |
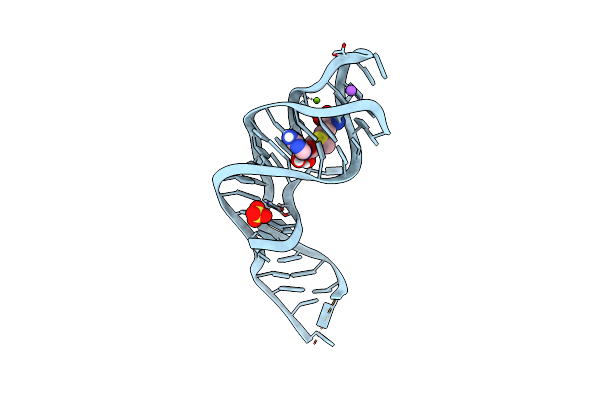 |
Organism: Candidatus pelagibacter ubique htcc1062
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2018-07-04 Classification: RNA Ligands: SO4, NA, SAM, MG |

