Search Count: 1,626
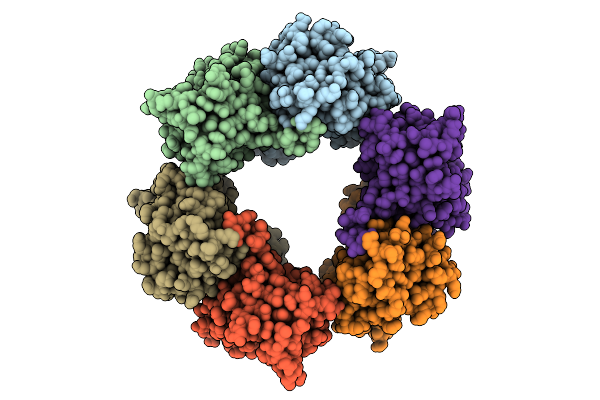 |
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In The Absence Of Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN |
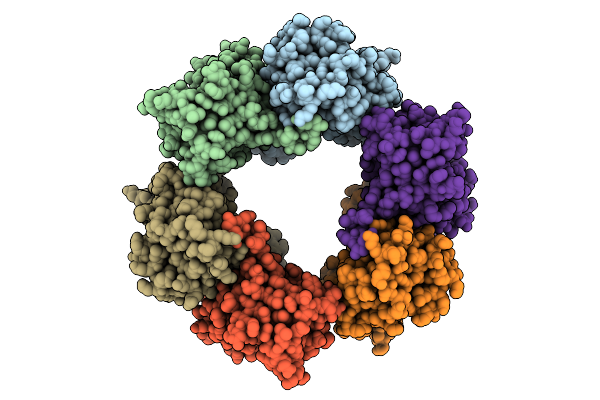 |
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In Complex With Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
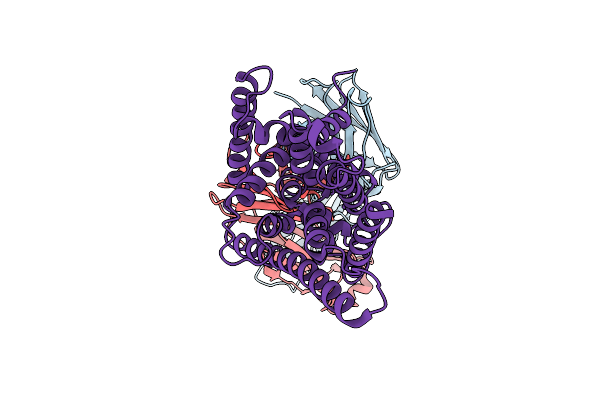 |
Cryo-Em Structure Of Candida Albicans Fluoride Channel Fex In Complex With Fab Fragment
Organism: Homo sapiens, Hiv-1 m:b_mn, Candida albicans sc5314
Method: ELECTRON MICROSCOPY Resolution:4.05 Å Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: PY1, CL |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: CL, A1B7R, SO4 |
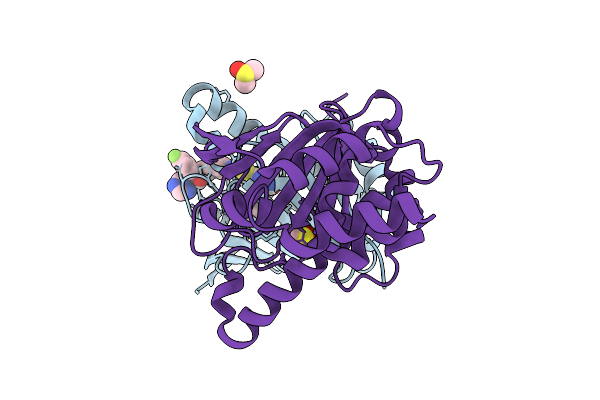 |
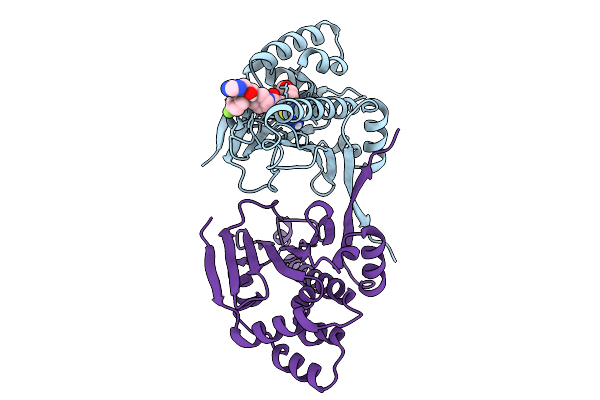
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2025-09-17 Classification: CHAPERONE Ligands: A1A27, DMS, CL |
 |
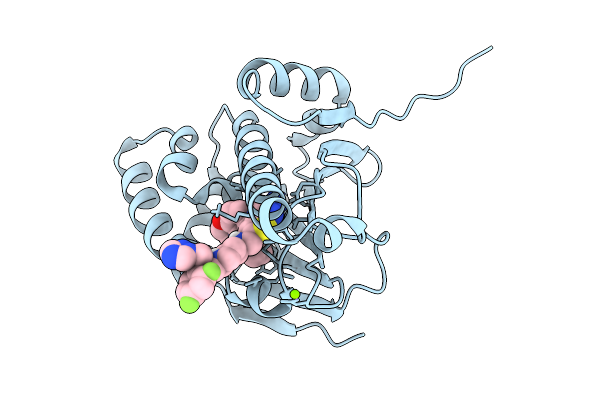
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A28 |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A26, MG |
 |
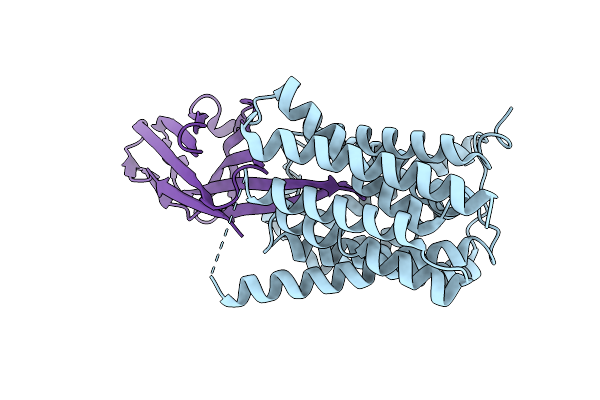
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A24, MG |
 |
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody.
Organism: Lama glama, Candida albicans
Method: ELECTRON MICROSCOPY Resolution:3.00 Å Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN |
 |
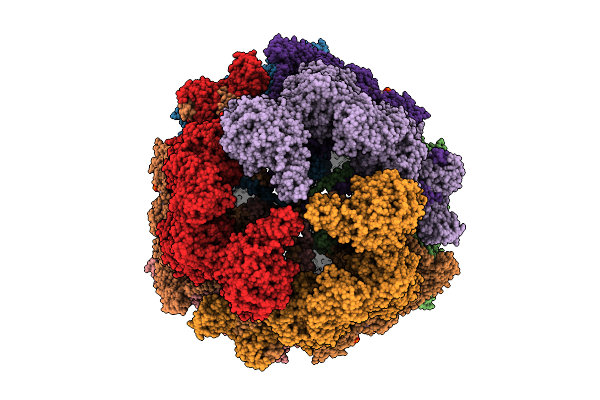
Cryo-Em Structure Of Candida Albicans Vrg4 Bound To An Inhibitory Nanobody And Gdp-Mannose.
Organism: Candida albicans, Lama glama
Method: ELECTRON MICROSCOPY Resolution:3.40 Å Release Date: 2025-09-03 Classification: TRANSPORT PROTEIN Ligands: GDD |
 |
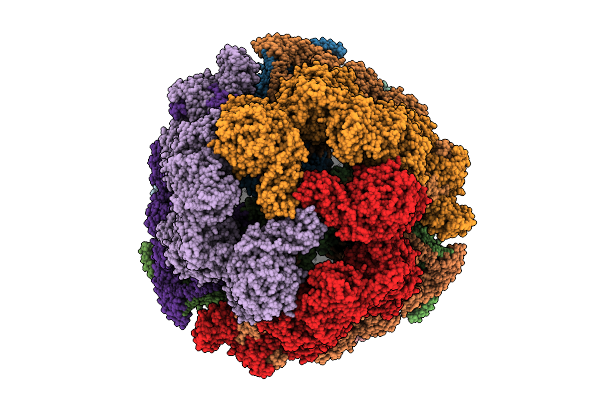
Atomic Model Of Candida Albicans Fatty Acid Synthase (Fas) In Complex With Palmitoyl-Coa (In Vitro Binding)
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN, PKZ |
 |
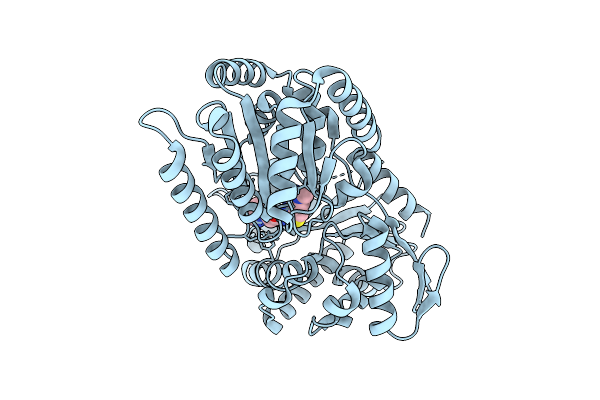
Atomic Model Of Ketoacyl Reductase Domain And 4 Helical Bundle Of Candida Albicans Fatty Acid Synthase (Fas) In Complex With Palmitoyl-Coa (In Vivo Binding)
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-08-20 Classification: BIOSYNTHETIC PROTEIN Ligands: FMN |
 |
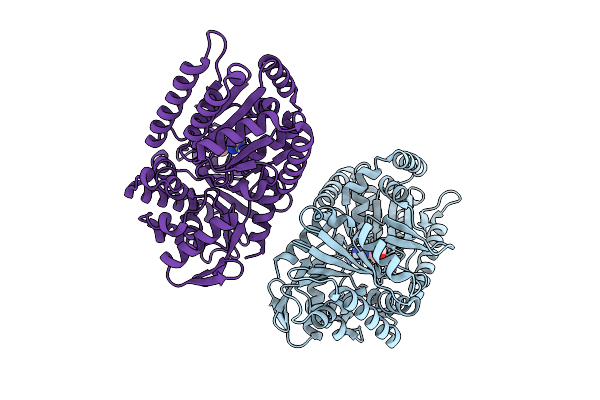
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With 4456
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:3.35 Å Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYI |
 |
Structure Of Candida Albicans Trehalose-6-Phosphate Synthase In Complex With Sj6675
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:3.50 Å Release Date: 2025-08-20 Classification: TRANSFERASE Ligands: A1BYV |
 |
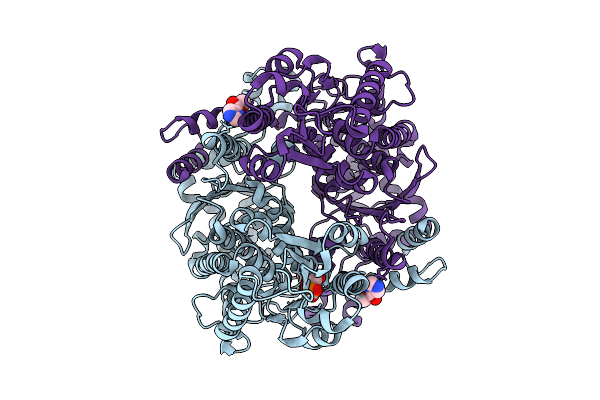
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.53 Å Release Date: 2025-07-30 Classification: ISOMERASE Ligands: PA5, A1IHU |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.49 Å Release Date: 2025-07-30 Classification: ISOMERASE Ligands: A1IIB, PA5, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IH9, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerae (Capgi) In Complex With Fragments
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IIA, A1IH8, CL |
 |
The Structure Of Candida Albicans Phosphoglucose Isomerase In Complex With A Fragment Binder
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.51 Å Release Date: 2025-07-23 Classification: ISOMERASE Ligands: PA5, A1IIC |

