Search Count: 6,290
All
Selected
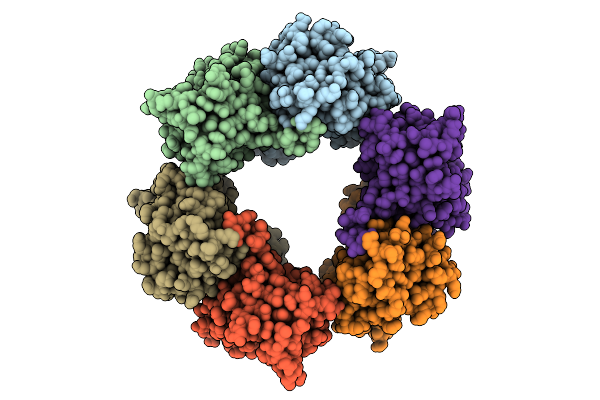 |
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In The Absence Of Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN |
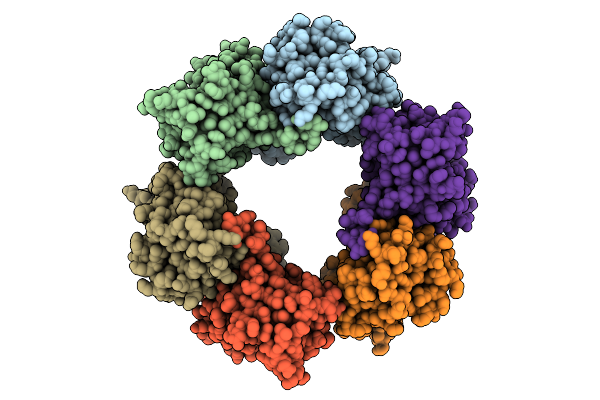 |
Cryo-Em Structure Of Candida Albicans Ph Regulated Antigen 1 (Pra1) Protein In Complex With Zn2+
Organism: Candida albicans
Method: ELECTRON MICROSCOPY Release Date: 2025-12-10 Classification: METAL BINDING PROTEIN Ligands: ZN |
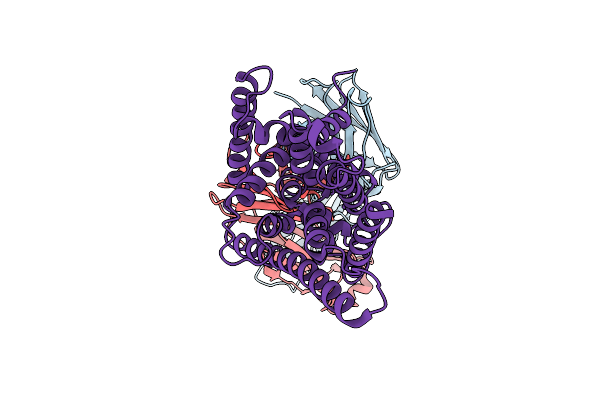 |
Cryo-Em Structure Of Candida Albicans Fluoride Channel Fex In Complex With Fab Fragment
Organism: Homo sapiens, Hiv-1 m:b_mn, Candida albicans sc5314
Method: ELECTRON MICROSCOPY Resolution:4.05 Å Release Date: 2025-12-03 Classification: MEMBRANE PROTEIN |
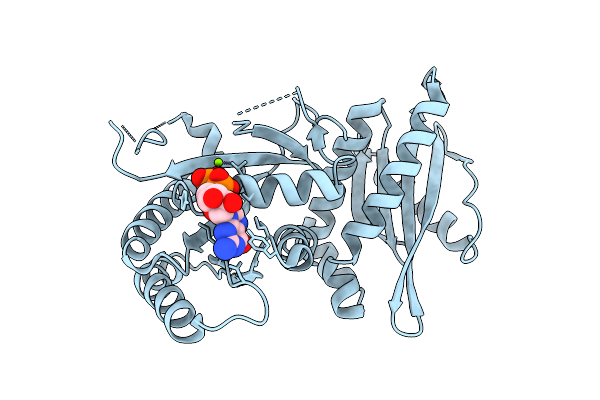 |
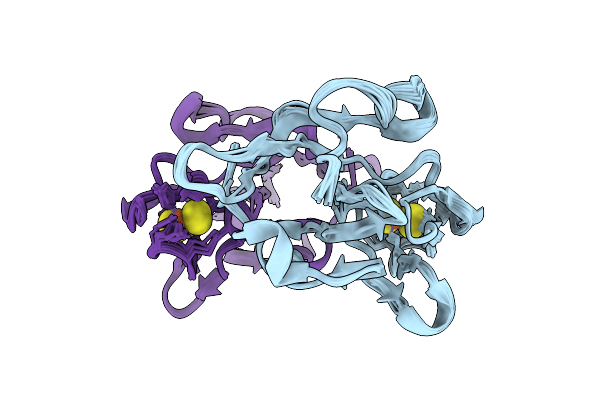
Crystal Structure Of Monomeric Rag-Like Small Gtpase From Asgard Lokiarchaeota (Lokiragm) In Complex With Gdp
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2025-11-26 Classification: HYDROLASE Ligands: GDP, MG |
 |
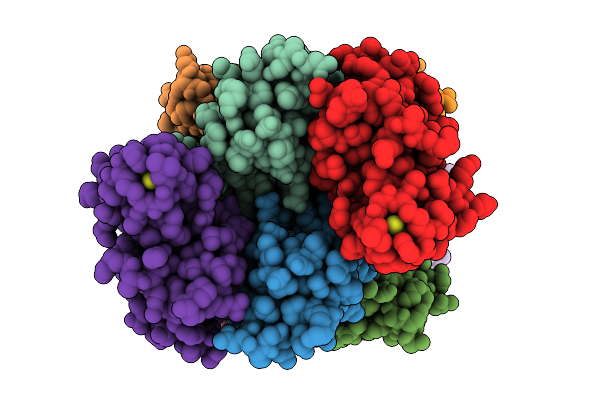
Soluble Domain Of Kustd1480, A Rieske Iron-Sulfur Cluster Protein From Kuenenia Stuttgartiensis
Organism: Candidatus kuenenia sp.
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2025-11-26 Classification: ELECTRON TRANSPORT Ligands: FES |
 |
Soluble Domain Of Kuste4569, A Rieske Iron-Sulfur Cluster Protein From Kuenenia Stuttgartiensis
Organism: Candidatus kuenenia sp.
Method: X-RAY DIFFRACTION Resolution:2.80 Å Release Date: 2025-11-26 Classification: ELECTRON TRANSPORT Ligands: FES, SO4 |
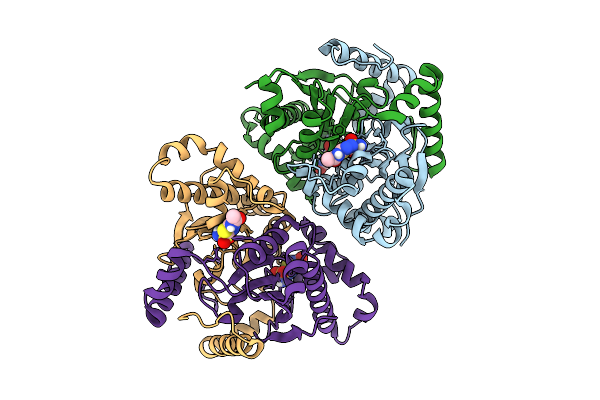 |
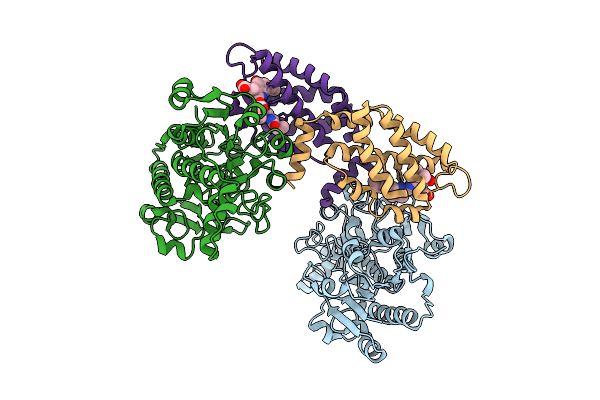
Organism: Candida parapsilosis
Method: X-RAY DIFFRACTION Resolution:2.70 Å Release Date: 2025-11-26 Classification: LYASE Ligands: ZN, AZM |
 |
Structure Of The Recombinant Structure Of The Subunit Of Allophycocyanin (Apc) And The Formate Dehydrogenase (Fdh)
Organism: Candida boidinii, Picosynechococcus sp. pcc 7003
Method: X-RAY DIFFRACTION Resolution:2.19 Å Release Date: 2025-11-05 Classification: PHOTOSYNTHESIS Ligands: CYC |
 |
Organism: Candidatus heimdallarchaeota archaeon lc_3, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.50 Å Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Candidatus heimdallarchaeota archaeon lc_3, Synthetic construct
Method: ELECTRON MICROSCOPY Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Candidatus heimdallarchaeota archaeon lc_3, Synthetic construct
Method: ELECTRON MICROSCOPY Resolution:3.60 Å Release Date: 2025-10-29 Classification: DNA BINDING PROTEIN |
 |
Organism: Candidatus odinarchaeum yellowstonii
Method: X-RAY DIFFRACTION Resolution:3.58 Å Release Date: 2025-10-15 Classification: TRANSFERASE Ligands: CL |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.70 Å Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: PY1, CL |
 |
Organism: Candida albicans
Method: X-RAY DIFFRACTION Resolution:1.92 Å Release Date: 2025-10-08 Classification: TRANSFERASE/INHIBITOR Ligands: CL, A1B7R, SO4 |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.03 Å Release Date: 2025-09-17 Classification: CHAPERONE Ligands: A1A27, DMS, CL |
 |
Crystal Structure Of Homo Tetrameric Raga-Like Small Gtpase From Asgard Lokiarchaeota (Lokirago) In Complex With Gtp
Organism: Candidatus prometheoarchaeum syntrophicum
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2025-09-10 Classification: HYDROLASE Ligands: GTP, MG |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:2.09 Å Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A28 |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:1.78 Å Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A26, MG |
 |
Organism: Candida albicans sc5314
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-09-10 Classification: CHAPERONE Ligands: A1A24, MG |
 |
Structural Basis Of Substrate Promiscuity In The Archaeal Rna-Splicing Endonuclease From Candidatus Micrarchaeum Acidiphilum (Arman-2)
Organism: Candidatus micrarchaeum acidiphilum arman-2, Archaeoglobus fulgidus
Method: X-RAY DIFFRACTION Resolution:1.80 Å Release Date: 2025-09-10 Classification: SPLICING/RNA Ligands: ACY, A23 |

