Search Count: 59
 |
Crystal Structure Of 2-Dehydro-3- Deoxygluconokinase (Ec 2.7.1.45) (Tm0067) From Thermotoga Maritima At 2.05 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2006-02-14 Classification: TRANSFERASE Ligands: NI, CA |
 |
Crystal Structure Of A Glutathione S-Transferase (Atu5508) From Agrobacterium Tumefaciens Str. C58 At 2.00 A Resolution
Organism: Agrobacterium tumefaciens
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2006-02-14 Classification: TRANSFERASE Ligands: SCN |
 |
Crystal Structure Of Glycerate Kinase (Ec 2.7.1.31) (Tm1585) From Thermotoga Maritima At 2.70 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.53 Å Release Date: 2005-11-08 Classification: TRANSFERASE |
 |
Crystal Structure Of Transcriptional Regulator, Tetr Family (Tm1030) From Thermotoga Maritima At 2.30 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2005-07-26 Classification: TRANSCRIPTION REGULATOR Ligands: UNL |
 |
Crystal Structure Of An Atypical Cyclophilin (Peptidylprolyl Cis-Trans Isomerase) (Tm1367) From Thermotoga Maritima At 1.90 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2005-07-26 Classification: UNKNOWN FUNCTION Ligands: 1PE, NI |
 |
Crystal Structure Of The Apbe Protein (Tm1553) From Thermotoga Maritima Msb8 At 1.58 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.58 Å Release Date: 2005-03-22 Classification: BIOSYNTHETIC PROTEIN Ligands: UNL, MRD, MPD |
 |
Crystal Structure Of Gtp Binding Regulator (Tm1622) From Thermotoga Maritima At 1.75 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2005-03-15 Classification: SIGNALING PROTEIN Ligands: AZI, GOL |
 |
Crystal Structure Of Putative 2-Phosphosulfolactate Phosphatase (15026306) From Clostridium Acetobutylicum At 2.6 A Resolution
Organism: Clostridium acetobutylicum
Method: X-RAY DIFFRACTION Resolution:2.49 Å Release Date: 2005-02-15 Classification: HYDROLASE Ligands: MG, 3SL |
 |
Crystal Structure Of Acireductone Dioxygenase (13543033) From Mus Musculus At 2.06 A Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:2.06 Å Release Date: 2005-02-15 Classification: OXIDOREDUCTASE Ligands: NI, UNL, IPA |
 |
Crystal Structure Of Phosphoribosylformylglycinamidine Synthase, Purs Subunit (Ec 6.3.5.3) (Tm1244) From Thermotoga Maritima At 1.90 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-12-28 Classification: LIGASE |
 |
Crystal Structure Of A Virulence Factor (Cj0248) From Campylobacter Jejuni Subsp. Jejuni At 2.25 A Resolution
Organism: Campylobacter jejuni subsp. jejuni nctc 11168
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2004-12-28 Classification: UNKNOWN FUNCTION |
 |
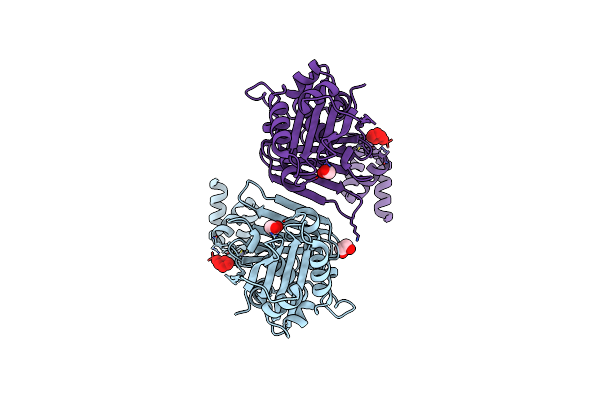
Crystal Structure Of A Putative Carbon Storage Regulator Protein (Csra, Pa0905) From Pseudomonas Aeruginosa At 2.05 A Resolution
Organism: Pseudomonas aeruginosa
Method: X-RAY DIFFRACTION Resolution:2.05 Å Release Date: 2004-12-14 Classification: RNA BINDING PROTEIN |
 |
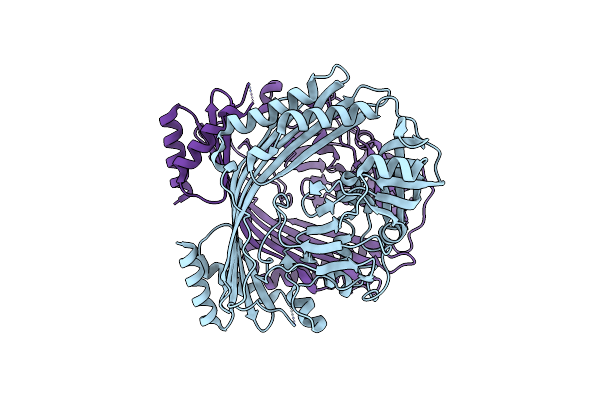
Crystal Structure Of 33 Kda Chaperonin (Heat Shock Protein 33 Homolog) (Hsp33) (Tm1394) From Thermotoga Maritima At 2.20 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2004-12-14 Classification: CHAPERONE Ligands: ZN, CL, UNL, EDO |
 |
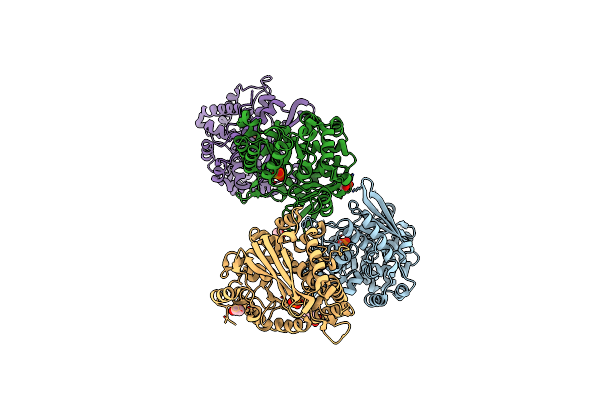
Crystal Structure Of A Putative Modulator Of A Dna Gyrase (Tm0727) From Thermotoga Maritima Msb8 At 1.95 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.95 Å Release Date: 2004-09-21 Classification: GENE REGULATION |
 |
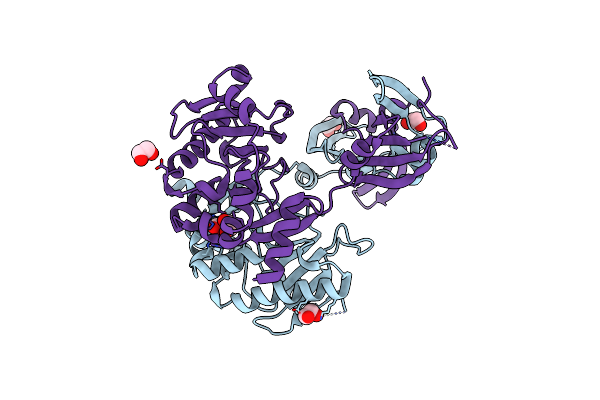
Crystal Structure Of A Putative Nicotinate Phosphoribosyltransferase (Yor209C, Npt1) From Saccharomyces Cerevisiae At 1.75 A Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.75 Å Release Date: 2004-08-24 Classification: TRANSFERASE Ligands: PO4, CL, MES, EDO |
 |
Crystal Structure Of Mrna Decapping Enzyme (Dcps) From Mus Musculus At 1.83 A Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.83 Å Release Date: 2004-08-24 Classification: RNA BINDING PROTEIN Ligands: EDO |
 |
Crystal Structure Of S-Adenosylmethionine Trna Ribosyltransferase (Tm0574) From Thermotoga Maritima At 2.00 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2004-08-17 Classification: ISOMERASE Ligands: UNL |
 |
Crystal Structure Of An Indigoidine Synthase A (Idga)-Like Protein (Tm1464) From Thermotoga Maritima Msb8 At 1.90 A Resolution
Organism: Thermotoga maritima
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-06-29 Classification: BIOSYNTHETIC PROTEIN Ligands: MN, UNL, EDO |
 |
Crystal Structure Of A Putative Serine Hydrolase (Ydr428C) From Saccharomyces Cerevisiae At 1.85 A Resolution
Organism: Saccharomyces cerevisiae
Method: X-RAY DIFFRACTION Resolution:1.85 Å Release Date: 2004-06-08 Classification: HYDROLASE Ligands: GOL, CL |
 |
Crystal Structure Of An Aig2-Like Protein (A2Ld1, Ggact, Mgc7867) From Mus Musculus At 1.90 A Resolution
Organism: Mus musculus
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2004-05-18 Classification: TRANSFERASE Ligands: FMT |

