Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
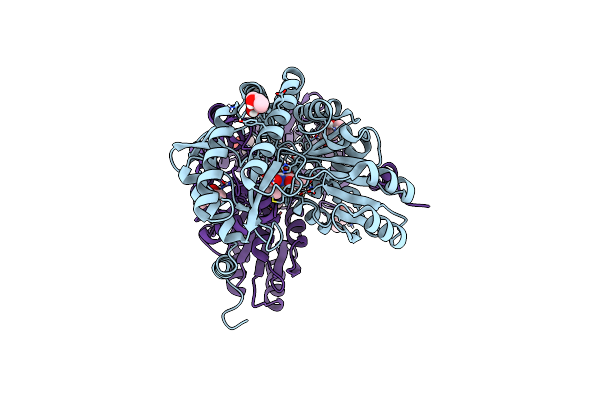 |
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Release Date: 2025-03-05 Classification: LIGASE Ligands: MPD, A1H4V |
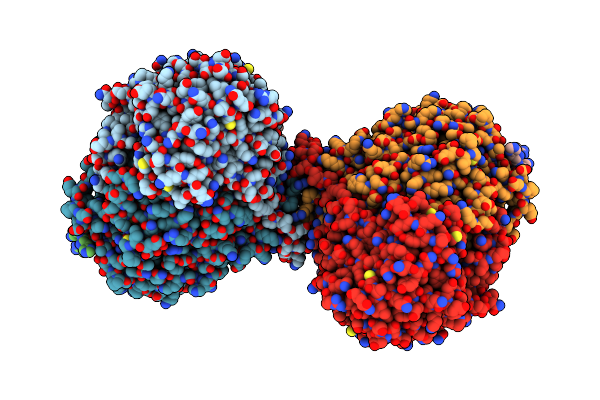 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae s288c , Saccharomyces cerevisiae, Saccharomyces cerevisiae s288c
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR Ligands: PLP, Z1T |
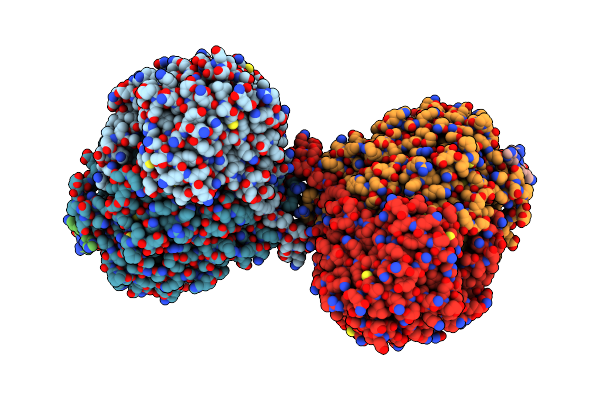 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae s288c, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR |
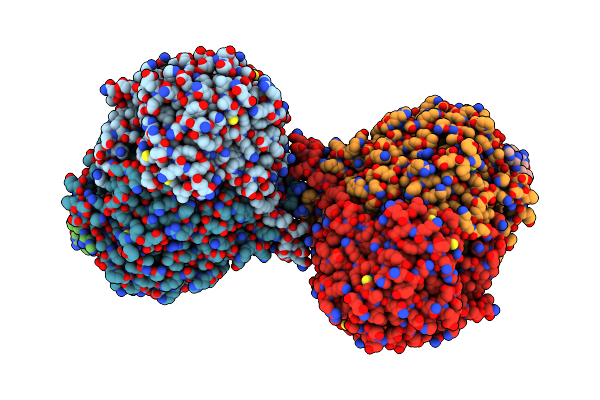 |
Organism: Arabidopsis thaliana, Saccharomyces cerevisiae
Method: ELECTRON MICROSCOPY Release Date: 2024-02-14 Classification: TRANSFERASE/INHIBITOR Ligands: PLP, Z1T |
 |
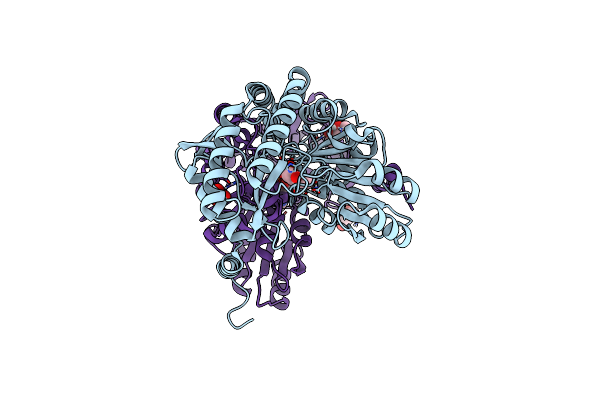
Crosslinked Crystal Structure Of The 8-Amino-7-Oxonanoate Synthase, Biof, And Benzene Sulfonyl Fluoride-Crypto Acyl Carrier Protein, Bsf-Acp
Organism: Escherichia coli
Method: X-RAY DIFFRACTION Resolution:2.30 Å Release Date: 2023-07-12 Classification: TRANSFERASE Ligands: PLP, EDO, SO4, SWC |
 |
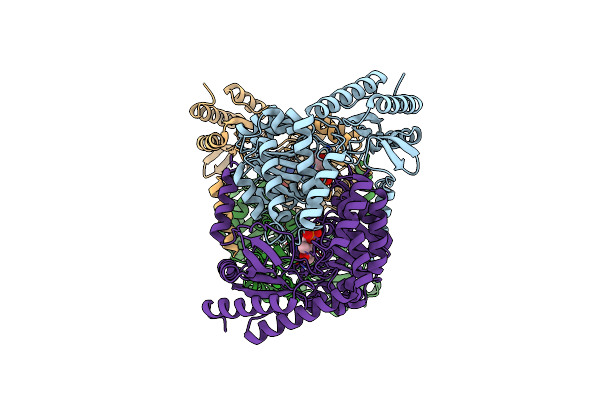
An Irreversible, Promiscuous And Highly Thermostable Claisen-Condensation Biocatalyst Drives The Synthesis Of Substituted Pyrroles
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:1.60 Å Release Date: 2022-09-21 Classification: SYNTHASE Ligands: PLP, MPD, SO4 |
 |
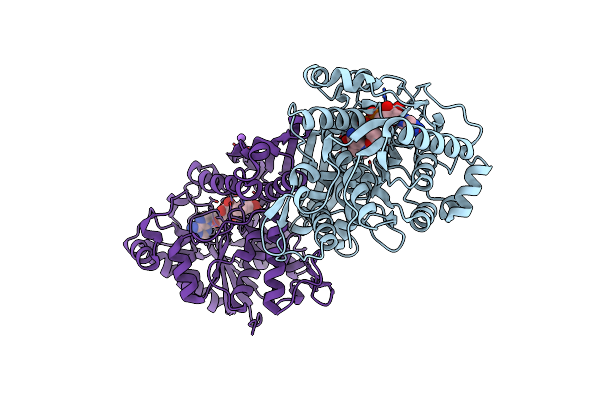
An Irreversible, Promiscuous And Highly Thermostable Claisen-Condensation Biocatalyst Drives The Synthesis Of Substituted Pyrroles
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-09-21 Classification: SYNTHASE Ligands: PLP, NA |
 |
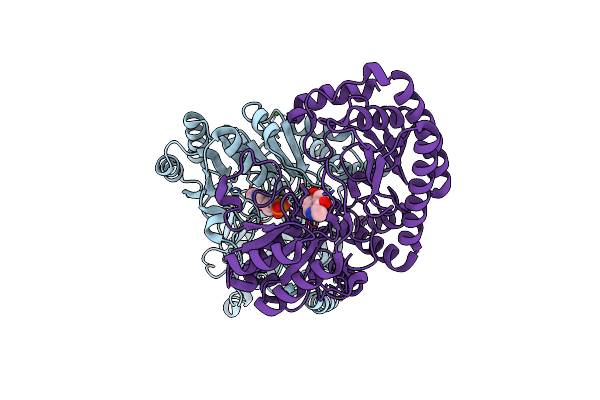
An Irreversible, Promiscuous And Highly Thermostable Claisen-Condensation Biocatalyst Drives The Synthesis Of Substituted Pyrroles
Organism: Thermus thermophilus
Method: X-RAY DIFFRACTION Resolution:2.60 Å Release Date: 2022-09-21 Classification: SYNTHASE Ligands: PLP |
 |
Organism: Bacteroides thetaiotaomicron
Method: X-RAY DIFFRACTION Resolution:2.00 Å Release Date: 2022-03-30 Classification: ISOMERASE Ligands: NAD, NA |
 |
Structure Of Plp Internal Aldimine Form Of Sphingopyxis Sp. Mta144 Fumi Protein
Organism: Sphingopyxis macrogoltabida
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-08-28 Classification: TRANSFERASE Ligands: PLP, MG |
 |
Structure Of Plp Internal Aldimine Form Of Sphingopyxis Sp. Mta144 Fumi Protein
Organism: Sphingopyxis macrogoltabida
Method: X-RAY DIFFRACTION Resolution:1.65 Å Release Date: 2019-08-28 Classification: TRANSFERASE Ligands: PLP, PMP, MG |
 |
Crystal Structure Of D-Phenylglycine Aninotransferase (D-Phgat) From Pseudomonas Stutzeri With Plp Internal Aldimine
Organism: Pseudomonas stutzeri
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2019-04-10 Classification: TRANSFERASE |
 |
Solution Structure Of The Apo Doublet Acyl Carrier Protein From Prodigiosin Biosynthesis
Organism: serratia sp. atcc 39006
Method: SOLUTION NMR Release Date: 2018-12-12 Classification: BIOSYNTHETIC PROTEIN |
 |
Crystal Structure Of The 6-Carboxyhexanoate-Coa Ligase (Biow)From Bacillus Subtilis In Complex With Coenzyme A
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.25 Å Release Date: 2017-03-08 Classification: LIGASE Ligands: PML, 2PE, COA |
 |
Crystal Structure Of The 6-Carboxyhexanoate-Coa Ligase (Biow)From Bacillus Subtilis In Complex With Amppnp
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.04 Å Release Date: 2016-11-16 Classification: LIGASE Ligands: PML, ANP, MG |
 |
Crystal Structure Of The 6-Carboxyhexanoate-Coa Ligase (Biow) From Bacillus Subtilis In Complex With A Pimeloyl-Adenylate
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.34 Å Release Date: 2016-11-16 Classification: LIGASE Ligands: WAQ, PPV, MG |
 |
Crystal Structure Of The 6-Carboxyhexanoate-Coa Ligase (Biow)From Bacillus Subtilis (Ptcl4 Derivative)
Organism: Bacillus subtilis
Method: X-RAY DIFFRACTION Resolution:2.44 Å Release Date: 2016-11-16 Classification: LIGASE Ligands: WAQ, PPV, MG, PT |
 |
Structure Of Burkholderia Pseudomallei K96243 Sphingosine-1-Phosphate Lyase Bpss2021
Organism: Burkholderia pseudomallei k96243
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2016-11-02 Classification: LYASE Ligands: PLP |
 |
Structure Of A C269A Mutant Of Propionaldehyde Dehydrogenase From The Clostridium Phytofermentans Fucose Utilisation Bacterial Microcompartment
Organism: Clostridium phytofermentans (strain atcc 700394 / dsm 18823 / isdg)
Method: X-RAY DIFFRACTION Resolution:1.77 Å Release Date: 2016-03-16 Classification: OXIDOREDUCTASE Ligands: SO4, COA, ACT |
 |
Structure Of His387Ala Mutant Of The Propionaldehyde Dehydrogenase From The Clostridium Phytofermentans Fucose Utilisation Bacterial Microcompartment
Organism: Clostridium phytofermentans
Method: X-RAY DIFFRACTION Resolution:2.08 Å Release Date: 2016-03-16 Classification: OXIDOREDUCTASE Ligands: SO4 |