Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
Planned Maintenance: Some services may turn out to be unavailable from 15th January, 2026 to 16th January, 2026. We apologize for the inconvenience!
 |
Organism: Norovirus, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR |
 |
Organism: Severe acute respiratory syndrome coronavirus 2, Synthetic construct
Method: X-RAY DIFFRACTION Release Date: 2025-12-31 Classification: VIRAL PROTEIN/INHIBITOR Ligands: SO4, NA |
 |
Minimal Puta Proline Dehydrogenase Domain (Design #2) Covalently Inactivated By (S)-But-3-Yn-2-Ylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.68 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE Ligands: A1BC6, FMT |
 |
Proline Utilization A (Puta) From Sinorhizobium Meliloti Inactivated By N-Propargylglycine
Organism: Sinorhizobium meliloti
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2025-02-05 Classification: OXIDOREDUCTASE/INHIBITOR Ligands: P5F, NAD, SO4, MG, FMT, PEG, PG4, PGE |
 |
Structure Of Norovirus (Hu/Gii.4/Sydney/Nsw0514/2012/Au) Protease In The Ligand-Free State
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: X-RAY DIFFRACTION Resolution:2.79 Å Release Date: 2024-01-31 Classification: VIRAL PROTEIN |
 |
Structure Of Norovirus (Hu/Gii.4/Sydney/Nsw0514/2012/Au) Protease Bound To Inhibitor Nv-004
Organism: Norovirus hu/gii.4/sydney/nsw0514/2012/au
Method: X-RAY DIFFRACTION Resolution:1.84 Å Release Date: 2023-12-20 Classification: VIRAL PROTEIN/INHIBITOR Ligands: GOL, ACT, FHR |
 |
Structure Of Proline Utilization A With 1,3-Dithiolane-2-Carboxylate Bound In The Proline Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.63 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJD, FAD, NAD, SO4, MG, PGE, PEG, PG4 |
 |
Structure Of Proline Utilization A With The Fad Covalently-Modified By 1,3-Dithiolane
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJJ, MG, NAD, SO4, PGE, PEG, PG4 |
 |
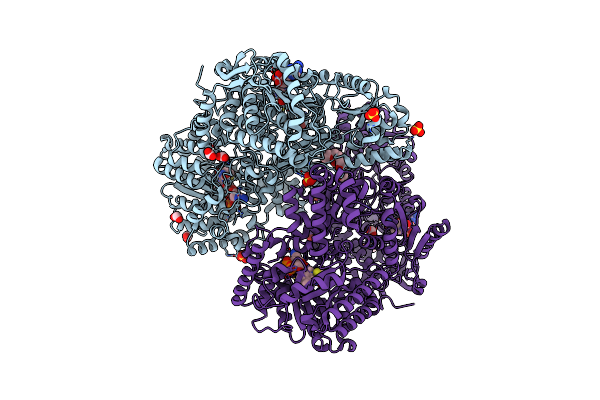
Structure Of Proline Utilization A With Tetrahydrothiophene-2-Carboxylate Bound In The Proline Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.52 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJM, UJP, PGE, FAD, NAD, SO4, MG, PEG |
 |
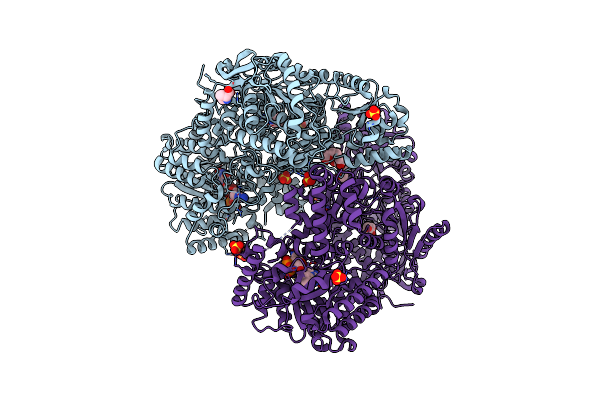
Structure Of Proline Utilization A With The Fad Covalently Modified By Tetrahydrothiophene
Organism: Sinorhizobium meliloti sm11
Method: X-RAY DIFFRACTION Resolution:1.90 Å Release Date: 2021-09-29 Classification: OXIDOREDUCTASE Ligands: UJG, NAI, SO4, PGE, FMT, MG, PEG |
 |
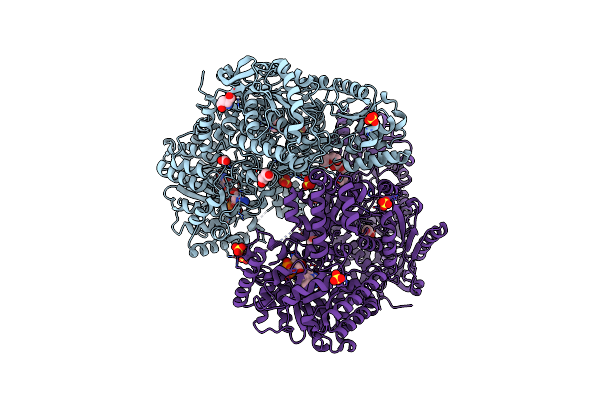
Structure Of Proline Utilization A With D-Proline Bound In The L-Glutamate-Gamma-Semialdehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.56 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: FAD, SO4, DPR, PGE |
 |
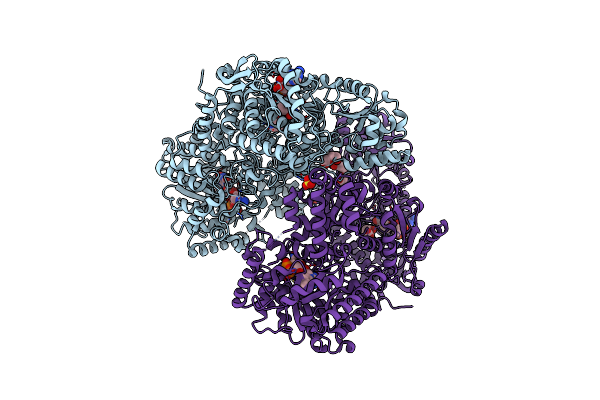
Structure Of Proline Utilization A With Trans-4-Hydroxy-D-Proline Bound In The L-Glutamate-Gamma-Semialdehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.41 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: FAD, FMT, PEG, UY7, SO4, PGE |
 |
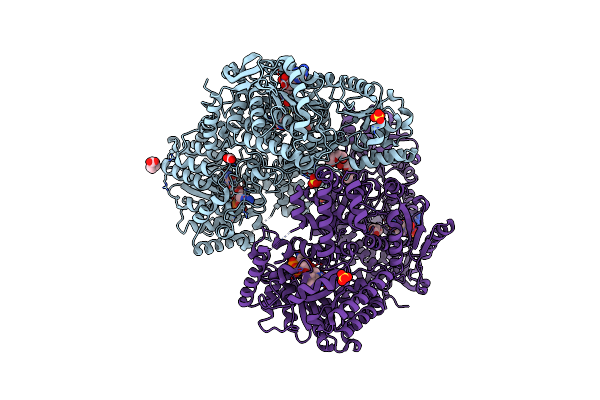
Structure Of Proline Utilization A With Cis-4-Hydroxy-D-Proline Bound In The L-Glutamate-Gamma-Semialdehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.46 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: FAD, PEG, NAD, SO4, MG, UYA, PGE |
 |
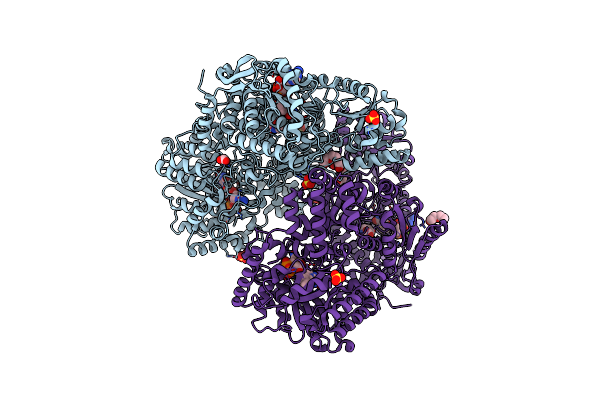
Structure Of Proline Utilization A With L-Proline Bound In The L-Glutamate-Gamma-Semialdehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.44 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: NAI, PEG, FMT, FDA, SO4, MG, PRO, PGE |
 |
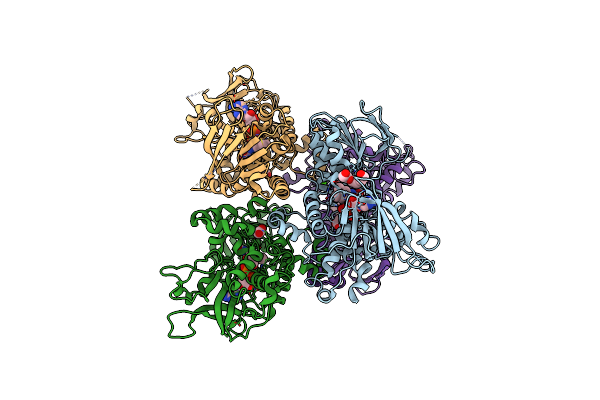
Structure Of Proline Utilization A With Trans-4-Hydroxy-L-Proline Bound In The L-Glutamate-Gamma-Semialdehyde Dehydrogenase Active Site
Organism: Sinorhizobium meliloti (strain sm11)
Method: X-RAY DIFFRACTION Resolution:1.54 Å Release Date: 2020-12-30 Classification: OXIDOREDUCTASE Ligands: FAD, HYP, PGE, FMT, NAD, SO4, MG, PEG |
 |
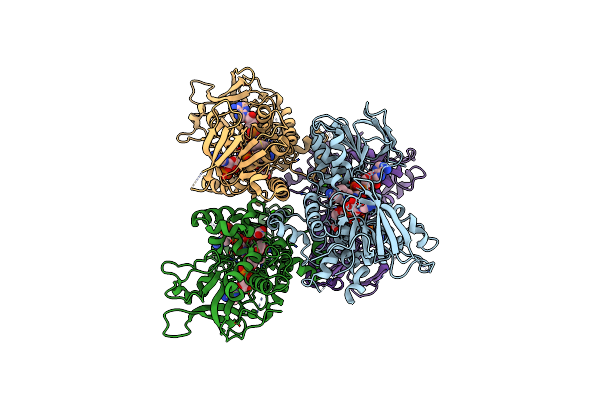
Structure Of The M101A Variant Of The Sida Ornithine Hydroxylase With The Fad In The "Out" Conformation
Organism: Neosartorya fumigata
Method: X-RAY DIFFRACTION Resolution:2.20 Å Release Date: 2020-12-02 Classification: OXIDOREDUCTASE Ligands: FAD, PGE, ACT, CA |
 |
Structure Of The M101A Variant Of The Sida Ornithine Hydroxylase Complexed With Nadp And The Fad In The "Out" Conformation
Organism: Neosartorya fumigata
Method: X-RAY DIFFRACTION Resolution:2.10 Å Release Date: 2020-12-02 Classification: OXIDOREDUCTASE Ligands: FAD, NAP, ACT, CA |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.50 Å Release Date: 2020-08-05 Classification: OXIDOREDUCTASE Ligands: NAD |
 |
Organism: Homo sapiens
Method: X-RAY DIFFRACTION Resolution:2.64 Å Release Date: 2020-08-05 Classification: OXIDOREDUCTASE Ligands: NAD |